Session Information
Date: Wednesday, November 8, 2017
Title: Systemic Lupus Erythematosus – Human Etiology and Pathogenesis II
Session Type: ACR Concurrent Abstract Session
Session Time: 11:00AM-12:30PM
Background/Purpose : Remarkable clinical and pathophysiological diversity complicate diagnosis, treatment and therapeutic development in systemic lupus erythematosus (SLE). This study used molecular phenotyping to identify more homogeneous subsets of lupus patients.
Methods: Plasma, serum and RNA serial or single samples (n=290) were collected from 184 SLE patients who met ACR classification. Disease activity was assessed by modified SELENA-SLEDAI. Immune pathways were evaluated by modular transcriptional analysis of Illumina Beadchip Microarray gene expression data. Plasma soluble mediators (n=32) and 13 antinuclear autoantibodies (anti-dsDNA, chromatin, ribosomal P, Sm, Sm/RNP, RNP, centromere B, Scl-70 and Jo-1) were assessed by multiplex, bead-based assay and ELISAs. Spearman correlations were used for univariate and multivariate analysis using R. Patients were clustered on transcriptional module scores and soluble mediators using Random Forest and tSNE.
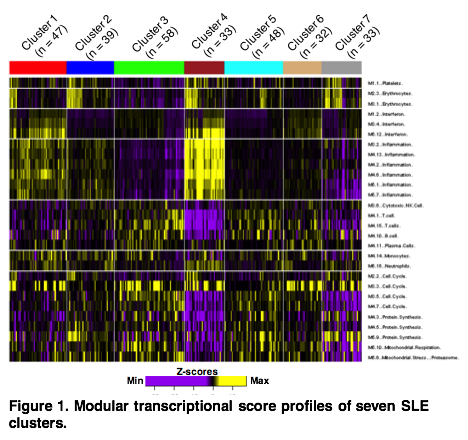
Results: We identified seven clusters of SLE patients with distinct molecular profiles. Cluster 4 had the highest inflammation and interferon (IFN) signatures, followed by Cluster 1. These clusters differed in IFN, T cell and inflammation module scores. Other clusters showed no elevation of IFN nor inflammation modules, but were distinguished by monocyte, neutrophil, plasma cell and T cell module scores. All three IFN expression modules strongly correlated with circulating IFN-related mediators, including IFNa, IFNg, IP10, MCP1, MIG, MIP1a, and MIP1b. The clusters with high IFN and inflammation scores had elevated IL-10, as did Cluster 6, which had the highest plasma cell module score and had only moderate IFN and inflammation module scores. Clinically, Clusters 1 and 4 were similar, displaying the highest SLEDAI scores, as well as increased rates of low complement, DNA binding antibodies, proteinuria and hematuria. Cluster 2 showed slight elevations in IFN and inflammation modules, but had low SLEDAI scores and reduced prevalence of low complement and DNA binding antibodies. Clusters 3, 5, 6, and 7 had higher rates of rash, alopecia and arthritis.
Conclusion: Molecular profiles encompassing IFN, T cell, neutrophil, plasma cell, and inflammation signatures distinguish groups of SLE patients and reveal multiple potential pathways of clinical disease. These profiles can potentially contribute to clinical trial design and individualized treatment.
To cite this abstract in AMA style:
Lu R, Guthridge JM, Arriens C, Aberle T, Kamp S, Munroe ME, Gross T, DeJager W, Macwana S, Roberts VC, Apel S, Chen H, Chakravarty E, Thanou K, Merrill JT, James JA. Molecular Phenotypes Associated with Clinical Disease Activity in Adult Systemic Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2017; 69 (suppl 10). https://acrabstracts.org/abstract/molecular-phenotypes-associated-with-clinical-disease-activity-in-adult-systemic-lupus-erythematosus/. Accessed .« Back to 2017 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/molecular-phenotypes-associated-with-clinical-disease-activity-in-adult-systemic-lupus-erythematosus/