Session Information
Session Type: Poster Session D
Session Time: 1:00PM-3:00PM
Background/Purpose: Guselkumab (GUS), a human monoclonal antibody targeting the interleukin-23p19 subunit, demonstrated joint and skin efficacy in patients with active psoriatic arthritis (PsA) in the Phase III DISCOVER-1/-2 trials.1,2 Minimal disease activity (MDA), a multi-domain composite outcome, is a clinically relevant measure of therapeutic response in PsA.3 However, response dynamics and the effect of individual domains on achieving MDA are not well understood. The purpose of this analysis was to characterize response patterns in MDA domains over time and identify potential baseline (BL) response predictors using machine learning.
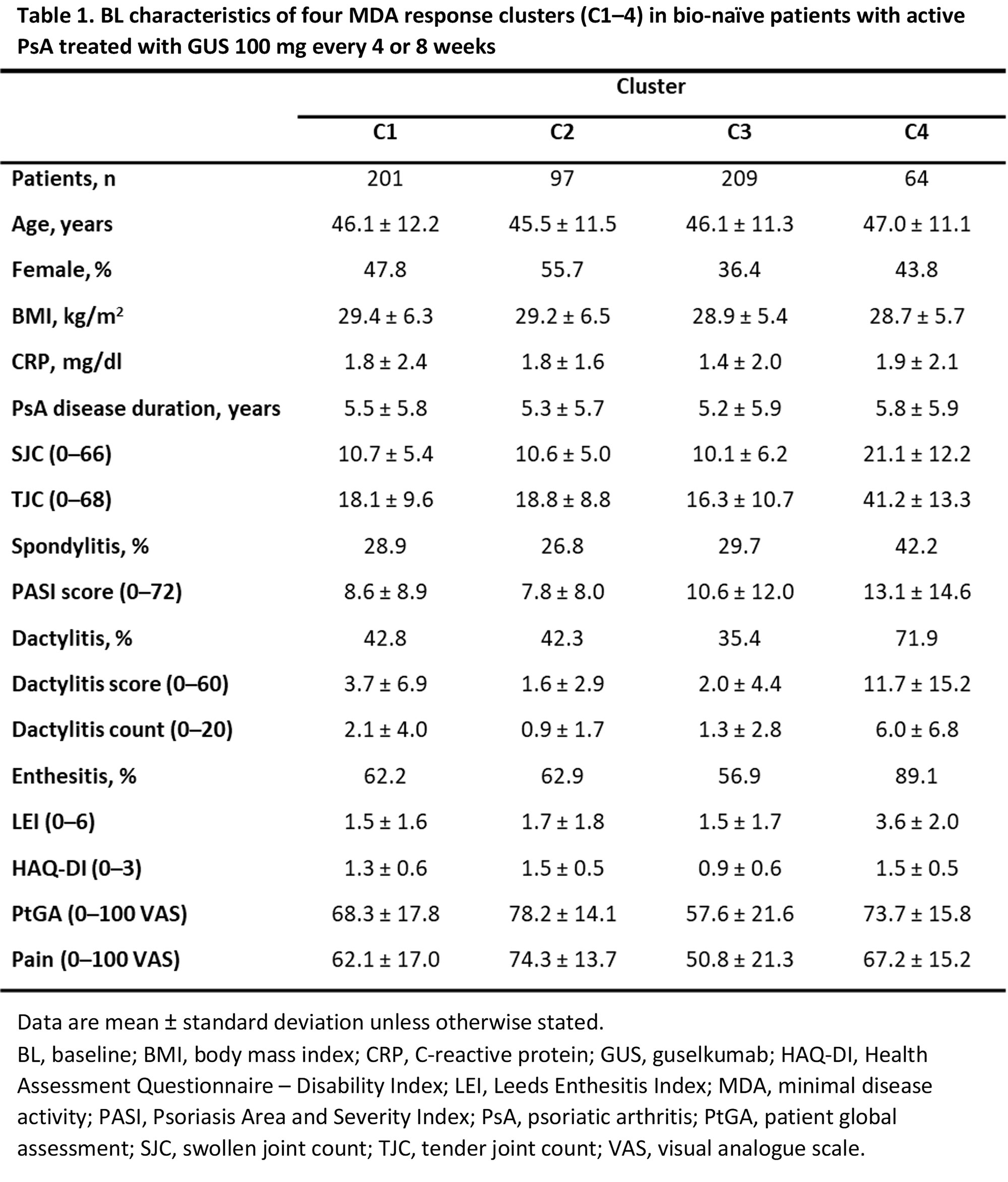
Methods: Data from bio-naïve patients with active PsA receiving GUS 100 mg every 4 or 8 weeks were pooled across DISCOVER-1/-2. Eligibility criteria are described elsewhere.1,2 Unsupervised machine learning using the time-series k-means clustering algorithm was performed to identify clusters according to MDA domain responses over 52 weeks. Individual MDA domain thresholds were swollen joint count (SJC), tender joint count (TJC), Psoriasis Area and Severity Index (PASI), and Leeds Enthesitis Index (LEI) each ≤1; patient global assessment (PtGA) visual analogue scale (VAS) ≤20; patient pain VAS ≤15; and Health Assessment Questionnaire-Disability Index (HAQ-DI) ≤0.5. BL characteristics were described for each cluster (Table 1). Missing data were not imputed.
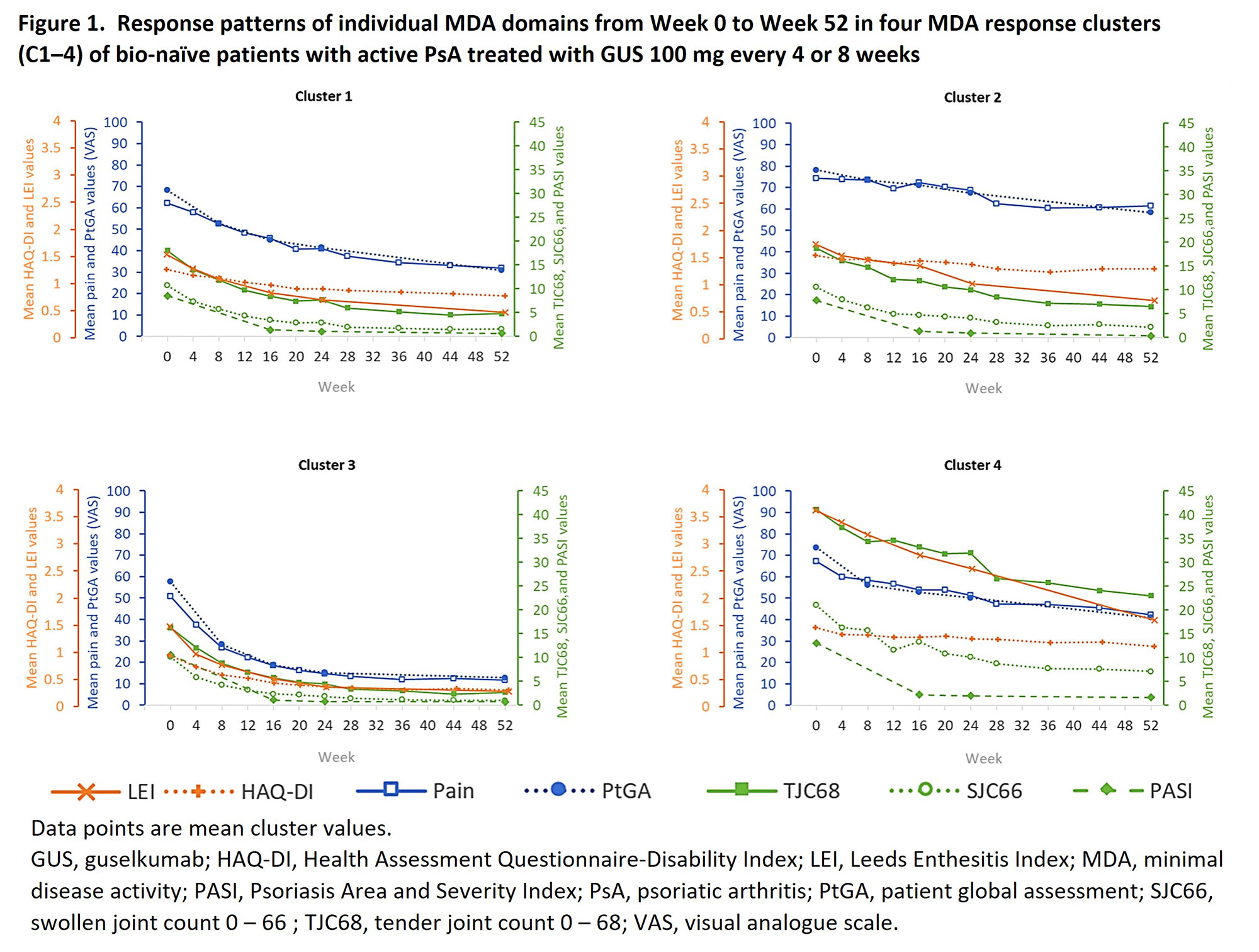
Results: This analysis included 571 of 669 patients receiving GUS and identified four distinct response clusters (C1–4; Table 1). Mean age and body mass index (BMI) were similar across clusters; C3 had a lower proportion of female patients. Relative to C3, a high burden of BL disease was observed in C4 across clinical measures and patient-reported outcomes (PROs), and across PROs only in C2. Through Week 52, MDA response rates were highest in C3 and lowest in C4, yet all clusters showed continuous improvement in mean values across all MDA domains (Figure 1). In C3, all individual domain thresholds were rapidly reached. C1, 2, and 4 met the PASI threshold and showed a substantial reduction in SJC, while other domains varied. In C1 and 2, improvement in clinical measures paralleled that of C3; however, PROs appeared to take longer to resolve. Responses were slowest in C4, though improvements were substantial given the high BL disease burden. Improvement in pain, PtGA, and HAQ-DI occurred earlier in C1 than C2.
Conclusion: Machine learning identified four clusters of GUS-treated patients with PsA based on differing response patterns in individual MDA domains. Response types may differ due to BL disease burden, especially in patients with higher pain, PtGA, and functional disability scores. These results offer an innovative, complementary approach to identifying treatment response patterns across diverse clusters of bio-naïve patients with active PsA, which may facilitate clinical decision-making.
References:
- Deodhar A et al. Lancet 2020; 395: 1115–1125.
- Mease PJ et al. Lancet 2020; 395: 1126–1136.
- Coates LC et al. Ann Rheum Dis 2010; 69: 48–53.
To cite this abstract in AMA style:
Zabotti A, Ohrndorf S, Tillett W, Neuhold M, van Speybroeck M, Theander E, Contré C, Sharaf M, Shawi M, Perate M, Kollmeier A, Richette P. Minimal Disease Activity Response Patterns in Bio-Naïve Patients Treated with Guselkumab: A Machine Learning Analysis [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/minimal-disease-activity-response-patterns-in-bio-naive-patients-treated-with-guselkumab-a-machine-learning-analysis/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/minimal-disease-activity-response-patterns-in-bio-naive-patients-treated-with-guselkumab-a-machine-learning-analysis/