Session Information
Date: Monday, October 22, 2018
Title: 4M090 ACR Abstract: Systemic Sclerosis & Rel D/O–Clinical II: Clinical Trials II (1875–1880)
Session Type: ACR Concurrent Abstract Session
Session Time: 2:30PM-4:00PM
Background/Purpose:
The SCOT (Scleroderma: Cyclophosphamide or Transplantation) trial (Sullivan K. et al, 2018) demonstrated the clinical benefit of hematopoietic stem cell transplant (HSCT) compared to cyclophosphamide (CYC) in dcSSc. We analyzed gene expression data from peripheral blood mononuclear cells (PBMCs) of SCOT participants to identify molecular changes and intrinsic gene expression subsets associated with treatment response.
Methods:
PBMC gene expression data were generated from 67 SCOT participants at baseline (36 CYC, 31 HSCT) and at 48/54 months (12 CYC, 14 HSCT). Significant differentially expressed genes (DEGs; False Discovery Rate <5%) were identified between baseline and 48/54 month samples using Significance Analysis of Microarrays and were annotated with Gene Ontology functional terms via g:Profiler. Participants who completed treatment protocol (Per-Protocol Population) were assigned to intrinsic gene expression subsets at baseline using a machine learning classifier based on elastic net multinomial regression previously trained and tested on five independent SSc cohorts. The relationship between intrinsic subsets and event-free survival (EFS) was analyzed at 54 months.
Results:
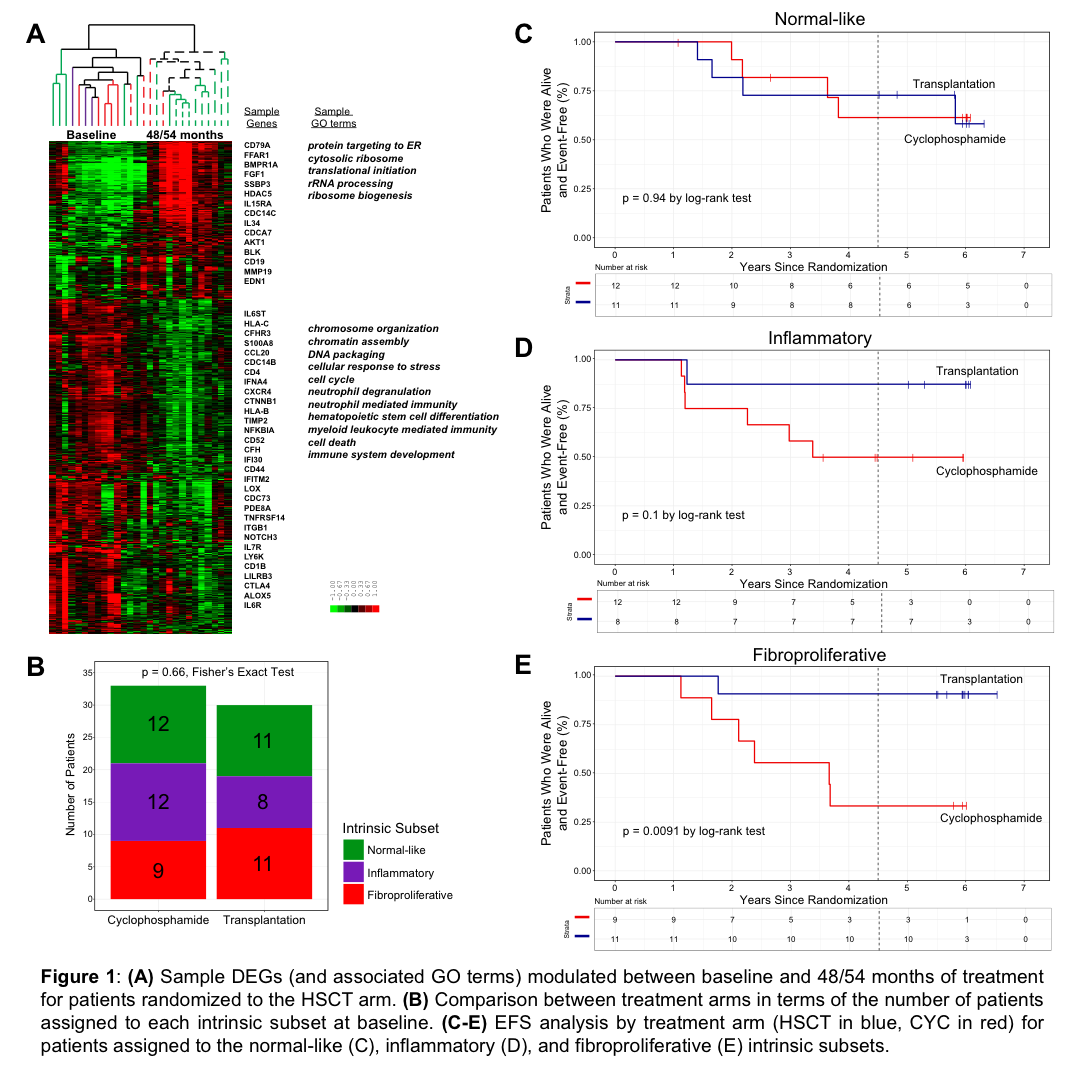
The participants included in the gene expression analyses were representative of the full cohort in the SCOT trial; there were no significant differences in sex, age, race, or baseline phenotypic measures. In the subset of participants with 48/54 month samples, we observed considerably more DEGs in HSCT arm (4788 genes) than in CYC arm (21 genes). Participants in HSCT showed a decrease in the expression of genes associated with cell proliferation and immune response and an increase in expression of genes associated with translation compared to baseline (Fig. 1A). Participants were assigned to intrinsic gene expression subsets at baseline (Fig. 1B) in both treatment arms. EFS did not differ between treatment arms for the participants assigned to the normal-like subset (p=0.94, Fig. 1C), consistent with the lack of active immune response in these participants. Participants assigned to the inflammatory subset trended towards improved chance of EFS in HSCT arm (p=0.1, Fig. 2D). Participants assigned to the fibroproliferative subset who received HSCT experienced significant improvement in EFS compared to fibroproliferative participants who received CYC (p=0.0091, Fig. 1E).
Conclusion:
The HSCT arm of the SCOT trial showed substantially larger changes in gene expression compared to CYC arm. Participants assigned to the normal-like subset did not show benefit from HSCT treatment over CYC. Importantly, participants assigned to the fibroproliferative subset, who tend not to improve on immunosuppressive therapy (e.g. MMF or abatacept), were the most likely to benefit from HSCT.
To cite this abstract in AMA style:
Franks J, Martyanov V, Wood TA, Crofford L, Keyes-Elstein L, Furst DE, Goldmuntz E, Mayes MD, McSweeney P, Nash R, Sullivan K, Whitfield ML. Machine Learning Classification of Peripheral Blood Gene Expression Identifies a Subset of Patients with Systemic Sclerosis Most Likely to Show Clinical Improvement in Response to Hematopoietic Stem Cell Transplant [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/machine-learning-classification-of-peripheral-blood-gene-expression-identifies-a-subset-of-patients-with-systemic-sclerosis-most-likely-to-show-clinical-improvement-in-response-to-hematopoietic-stem-c/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/machine-learning-classification-of-peripheral-blood-gene-expression-identifies-a-subset-of-patients-with-systemic-sclerosis-most-likely-to-show-clinical-improvement-in-response-to-hematopoietic-stem-c/