Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Familial Mediterranean Fever (FMF) is a monogenic autoinflammatory disease caused by mutations in the MEFV gene. Colchicine is the first-line treatment of FMF. Although it is an effective and safe option, %5 – %10 of FMF patients develop resistance to colchicine. To prevent attacks and subclinical inflammation, colchicine-resistant FMF is managed by elevating the colchicine dose or switching to IL-1 inhibitors. Predicting colchicine at the time of diagnosis would be beneficial for preventing amyloidosis and achieving better patient satisfaction by initiating a higher dose of colchicine earlier, preventing subclinical inflammation, and reducing the frequency of attacks. Therefore we aimed to develop machine learning algorithms for predicting colchicine resistance in patients with FMF.
Methods: We conducted an analysis of medical records from 1300 FMF patients, extracting various features including chest pain, presence of compound heterozygous mutations in MEFV gene, presence of M694V homozygous mutation, attacks per month before treatment, age of symptom onset, erysipelas-like rash associated with attacks, presence of arthritis with erythema, presence of recurrent arthritis, and colchicine resistance status.
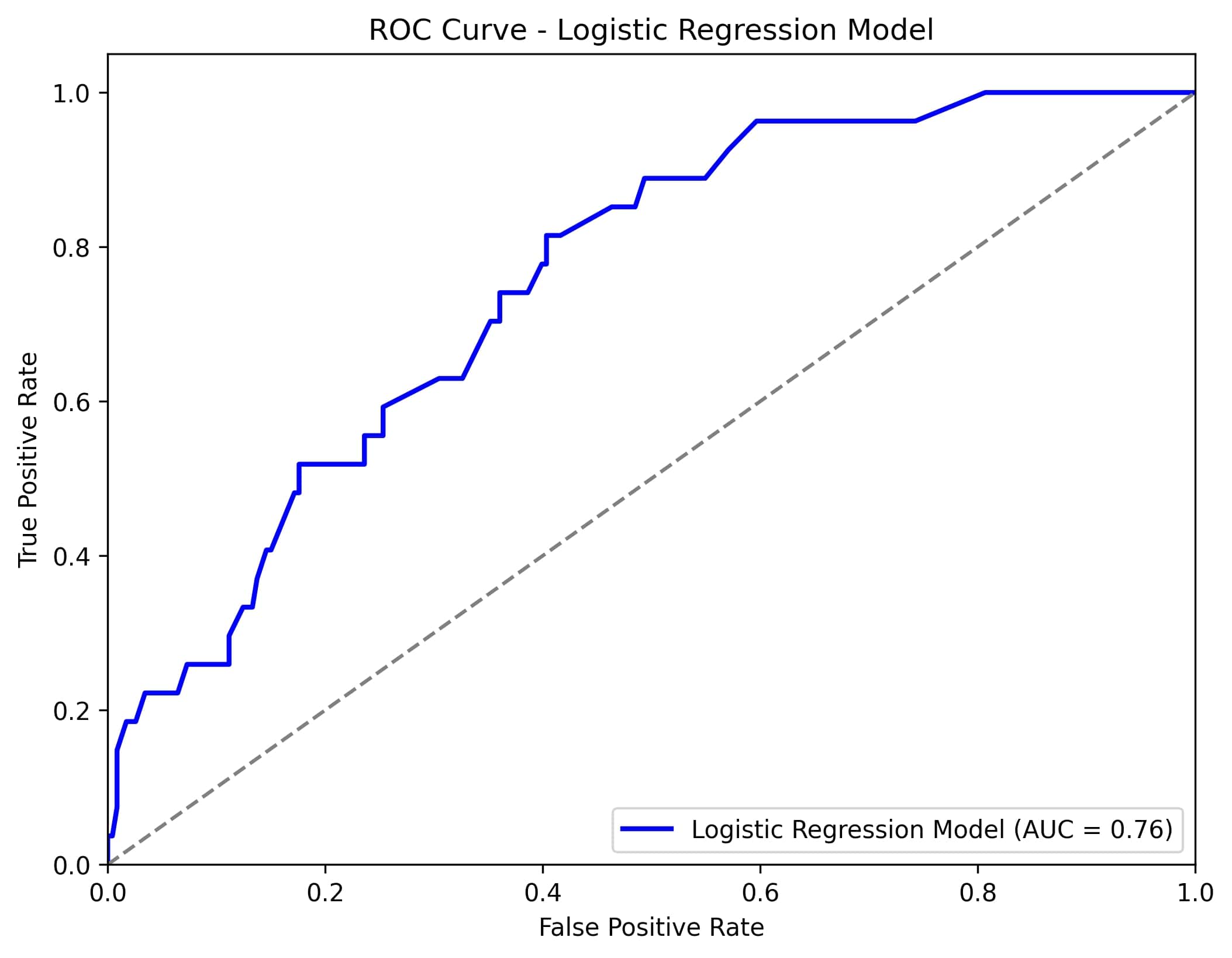
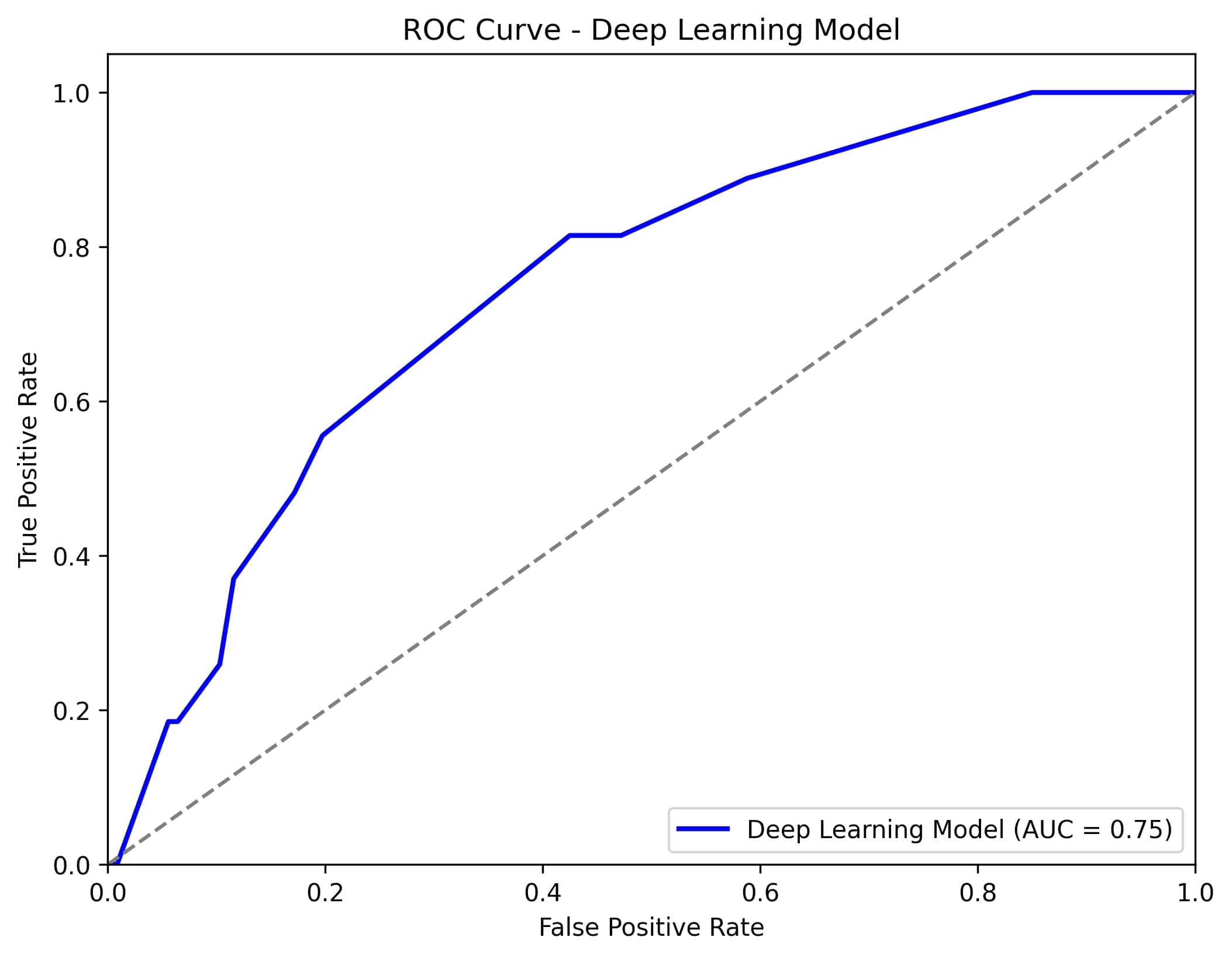
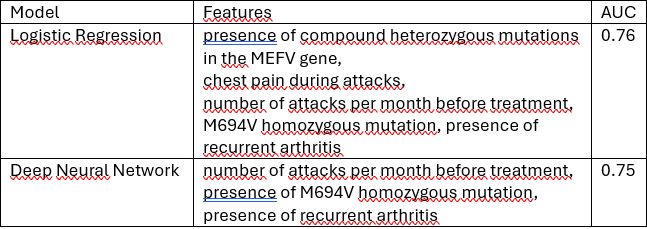
Results: The cohort included both pediatric and adult-onset FMF patients, with 113(8,6%) classified as colchicine resistant and 1187(91,4%) as colchicine-responsive. Logistic regression exhibited the best performance, utilizing features such as the presence of compound heterozygous mutations in the MEFV gene, chest pain during attacks, number of attacks per month before treatment, M694V homozygous mutation, and presence of recurrent arthritis, achieving an AUC of 0.76. (Figure 1) Deep neural network models also performed well, with the features of the number of attacks per month before treatment, the presence of M694V homozygous mutation, and the presence of recurrent arthritis, yielding an AUC of 0.75. (Figure 2)(Table 1)
Conclusion: Machine learning algorithms, offering a probability output for colchicine resistance risk, can function as sensitive and specific tests with adaptable thresholds. Enhancing AUC may require training deep learning models on larger datasets or incorporating additional features associated with colchicine resistance risk.
To cite this abstract in AMA style:
Öztürk A, Kucur M, Yagci L, Ugurlu S. Machine Learning Algorithms to Predict Colchicine Resistance in Familial Mediterranean Fever [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/machine-learning-algorithms-to-predict-colchicine-resistance-in-familial-mediterranean-fever/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/machine-learning-algorithms-to-predict-colchicine-resistance-in-familial-mediterranean-fever/