Session Information
Session Type: Abstract Submissions (ACR)
Background/Purpose
We have already reported that macrophage activation, and up-regulation of TGF-beta- and interferon-regulated genes are involved in progressive SSc-ILD. Micro-rnas (miRNA) are a class of small noncoding RNAs that control gene expression and are eventually involved in most biological processes. Therefore, we analyzed simultaneously mRNA and miRNA in the same prospective cohort of SSc-ILD patients in order to explore their complex network and the miRNA’s involvement in progressive lung disease.
Methods
Lung tissue was obtained by open lung biopsy in 22 consecutive SSc-ILD patients (11 diffuse and 10 limited cutaneous SSc patients; 5 controls). High-resolution computerized tomography (HRCT) was performed on baseline and 2-3 years after treatment that was based on lung histologic classification. Microarray analysis (mRNA and miRNA) was performed and the results correlated (Pearson’s) with changes in HRCT score (FibMax). MirConnX software was used to explore the gene regulatory network between mRNA and miRNA. The study was approved by the Institutional Review Boards from both universities (Brazil and USA).
Results
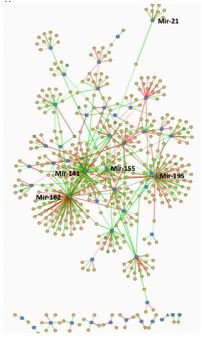
As already shown, despite treatment, most of SSc-ILD patients progressed based on delta FibMax (p< 0.01). Lung miRNA microarray analysis distinguished SSc-ILD from controls (FDR-corrected, q<0.25) with 185 miRNA genes (present in ³25%) that had significant differential expression. The mRNA microarray analysis also confirmed the altered expression of hundreds of genes in our SSc-ILD cohort and MirConnX simultaneous analysis of mRNA and miRNA showed 4 relevant miRNAs in the center of this multifaceted regulation (graphic): mir-182, mir-141, mir-155, and mir-195 (graphic). Pearson’s correlation between miRNAs and the top-50 most upregulated mRNAs in SSc-ILD compared to controls showed that mir-155 was strongly correlated (r>0.5) with 32/top-50 genes, such as IL13Ra2 (r=0.75) and TOP2A (r=0.68), mir-21 was the second most correlated (22/top-50 genes), followed by mir-195 (21/top-50 genes), mir-141 (22/top-50 genes), and mir-182 (19/top-50 genes). Mir-155 and mir-21 were also strongly negatively correlated with several top-downregulated mRNA genes, including IL12A, with r=-0.70. A heatmap of the most informative miRNAs showed mir-155 clustering together with mir-21, a known miRNA involved in idiopathic lung fibrosis; and both mir-155/mir-21 expression were strongly correlated (r2=0.6). Surprisingly, only 4 miRNAs were correlated to the delta FibMax: mir-155 (r=0,65), mir-182 (r=0.49), mir-27a (r=0.49), and mir-21 (r=0.47).
Conclusion
Mir-155 and mir-21 were the center of this complex altered gene expression in the lungs of SSc-ILD. More importantly, these miRNAs were key for the progression of the lung fibrosis based on a CT score. Together, anti-miRNA therapy might be a novel target to prevent progressive SSc-ILD.
Disclosure:
R. Christmann,
None;
P. D. Sampaio-Barros,
None;
C. Borges,
None;
C. Carvalho,
None;
R. A. Kairalla,
None;
E. R. Parra,
None;
G. Stifano,
None;
A. Spira,
None;
R. W. Simms,
None;
V. L. Capelozzi,
None;
R. Lafyatis,
None.
« Back to 2014 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/key-roles-for-mir-155-and-mir-21-in-progressive-lung-fibrosis-associated-with-systemic-sclerosis-ssc-ild/