Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: In cases where patients continue to experience active symptoms despite receiving maximum doses of nonsteroidal anti-inflammatory drug therapy in patients with axial spondyloarthritis (axSpA), TNF-α inhibitors (TNFi) are suggested as a second-line treatment. However, the response to TNFi is highly heterogeneous, with approximately 30% of patients being non-responsive. Yet, there is a notable absence of studies investigating prognostic markers for predicting the response to TNFi in individuals with axSpA, especially in genetic and transcriptional levels. We aimed to identify cell types and cell-type specific transcriptional markers associated with the response to TNFi in patients with axSpA.
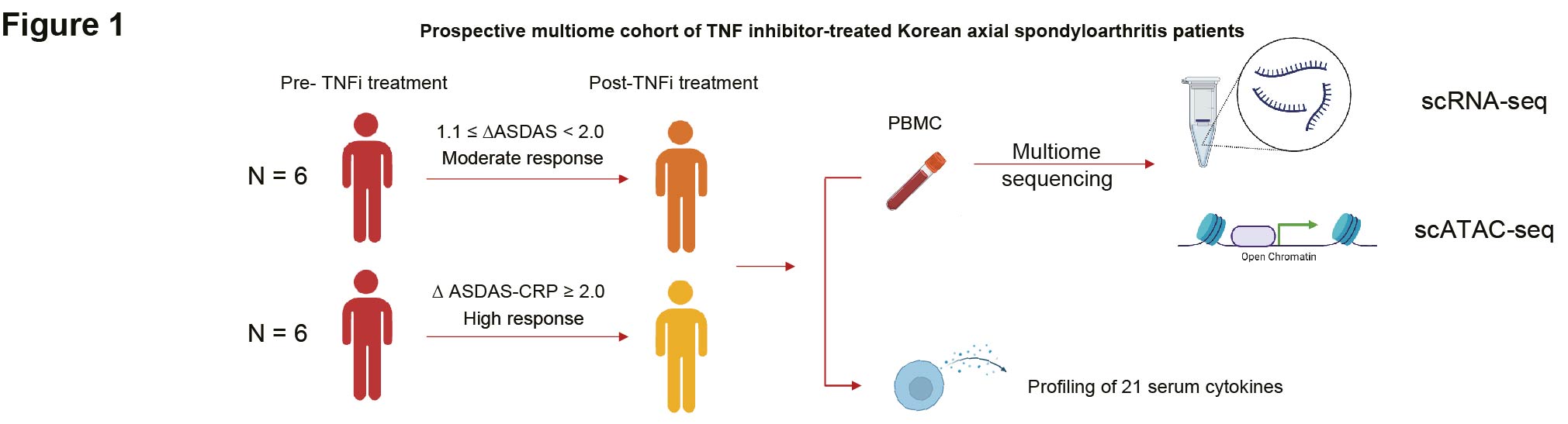
Methods: A prospective cohort of twelve biologics-naïve male patients with axSpA treated with TNFi at Samsung Medical Center underwent matched single-cell RNA sequencing (scRNA-seq) and single-cell assay for transposase accessible chromatin sequencing (scATAC-seq) on peripheral blood mononuclear cell (PBMC) samples collected before and three months after treatment (Figure 1). The cohort was categorized into high responders (patients meeting major improvement criteria, ∆ASDAS ≥ 2.0) and moderate responders (patients meeting clinically important improvement criteria, 1.1 ≤ ∆ASDAS < 2.0) based on the ASDAS response criteria.
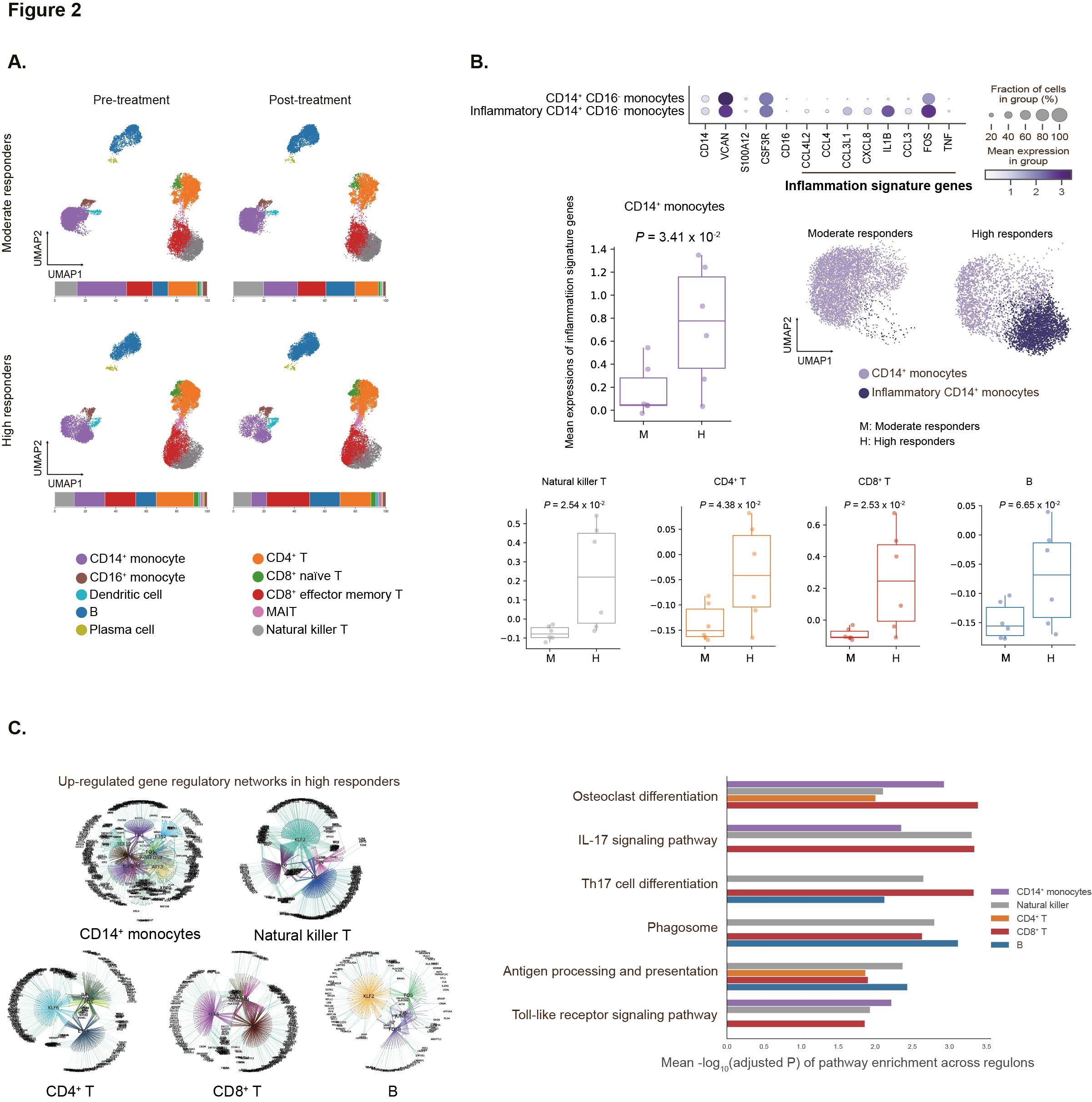
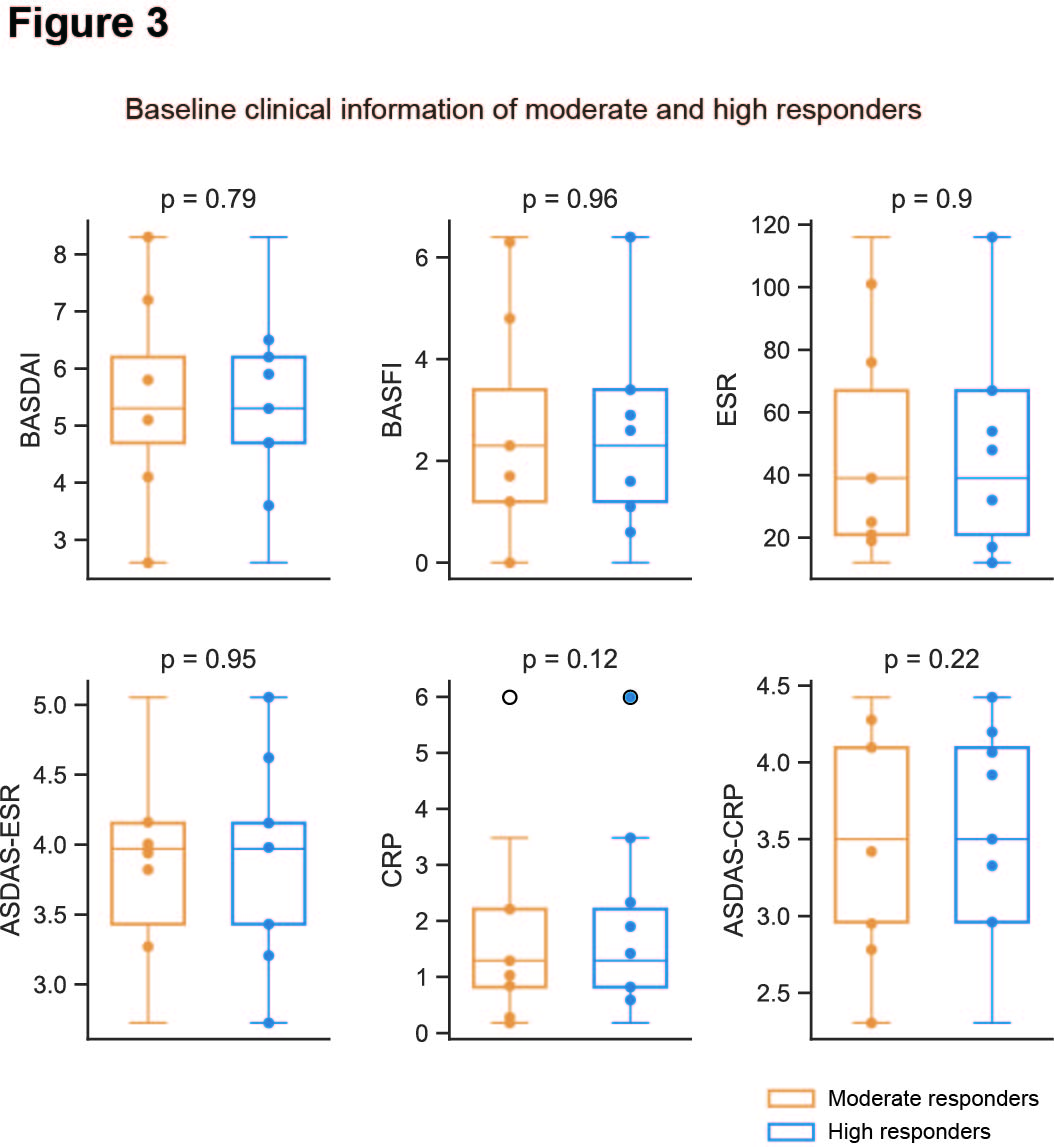
Results: We identified and characterized ten distinct cell types from PBMCs using the Leiden algorithm for 95,000 quality-filtered cells (Figure 2A). Among these, CD14+ monocytes, CD4+ T, CD8+ T, and B lymphocytes were the most abundant (Figure 2A). There were no significant differences in the proportions of these cell types between moderate and high responders at baseline. In high responders, however, a significantly higher fraction of CD14+ monocytes were in inflammatory states, characterized by the upregulation of eight inflammation signature genes (CCL42, CCL4, CXCL8, CCL3L1, IL1B, CCL3, FOS, and TNF), mainly involved in NFkB and IL17 signaling pathways (Figure 2B). Similar inflammatory patterns were observed in natural killer T, CD4+ T, CD8+ T, and B lymphocytes (Figure 2B). Using the SCENIC+ algorithm, enhancer-driven gene regulatory networks were inferred by integrating scRNA-seq and matched scATAC-seq data. Differential regulatory network analysis revealed that IL17 signaling and osteoclast differentiation pathways were significantly upregulated in CD14+ monocytes and T lymphocytes of high responders at baseline (Figure 2C). Interestingly, there were no significant differences in baseline clinical data (CRP, ASDAS-CRP, ESR, ASDAS-ESR, BASDAI, and BASFI) indicating symptom severity and inflammation between high and moderate responders (Figure 3).
Conclusion: This study provides insights into potential prognostic markers for TNFi response in axSpA patients, highlighting specific cell types and transcriptional markers associated with treatment outcomes. Identifying the inflammatory cell states of CD14+ monocyte as well as T and B lymphocytes and relevant biological pathways offer a foundation for further research and the development of personalized therapeutic strategies in the management of axSpA.
To cite this abstract in AMA style:
Lee S, Park J, Ignatova E, Kang S, Kim H, Lee J, Choi J, Cha H. Integration of Single-cell Multi-omics Profiling for Investigating Treatment Response to Tumor Necrosis Factor Alpha Inhibitors in Patients with Axial Spondyloarthritis [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/integration-of-single-cell-multi-omics-profiling-for-investigating-treatment-response-to-tumor-necrosis-factor-alpha-inhibitors-in-patients-with-axial-spondyloarthritis/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/integration-of-single-cell-multi-omics-profiling-for-investigating-treatment-response-to-tumor-necrosis-factor-alpha-inhibitors-in-patients-with-axial-spondyloarthritis/