Session Information
Date: Wednesday, November 13, 2019
Title: 6W021: RA – Treatments V: Switching & Tapering RA Medications (2906–2911)
Session Type: ACR Abstract Session
Session Time: 11:00AM-12:30PM
Background/Purpose: Efficiently identifying eligible patients is an important component of a successful clinical trial. Billing codes from electronic health record (EHR) data are commonly used to first screen for potential patients, followed by labor-intensive chart review to identify the eligible patients by trial criteria. The objective of this study was to test whether a machine learning screening algorithm (ML-screen) incorporating ICD codes and data extracted from notes using natural language processing (NLP), could improve the efficiency for identifying eligible patients for an ongoing clinical trial.
Methods: We studied EHR data used for a clinical recruitment study of rheumatoid arthritis (RA) and cardiovascular disease recruiting from a tertiary care center (TCC) and a community hospital (CH). The target population were RA patients, age >35, about to initiate a tumor necrosis factor inhibitor, and not on a statin. Prior to this study all patients with ≥1 RA ICD codes (RAICD) and age >35 years were selected for chart review. The CH and TCC data sets were both manually reviewed as gold standard labels including 642 and 2387 patients, respectively. All notes were processed with NLP to obtain the number of mentions for the concept of RA and inflammatory arthritis. Three groups of features were considered for the ML-screen (Table 1): (1) inclusion criteria features, e.g. RAICD; (2) exclusion criteria features, e.g. # of electronic prescriptions for a statin; (3) the total # ICD codes as a proxy for healthcare utilization. For the ML-screen we considered features within a 2-year timeframe prior to the chart review as well as all years prior. The ML-screen combined two ML methods, random forest (RF) and penalized logistic regression. The goal for the ML-screen was to reduce the number of patients requiring chart review without excluding potentially eligible patients. The ML-screen was compared to alternative approaches using RAICD ≥1, RAICD ≥2, and RAICD ≥1+exclusion criteria features. To test whether the ML-screen can be successfully ported to other institutions, we trained at TCC and applied at CH, and vice versa.
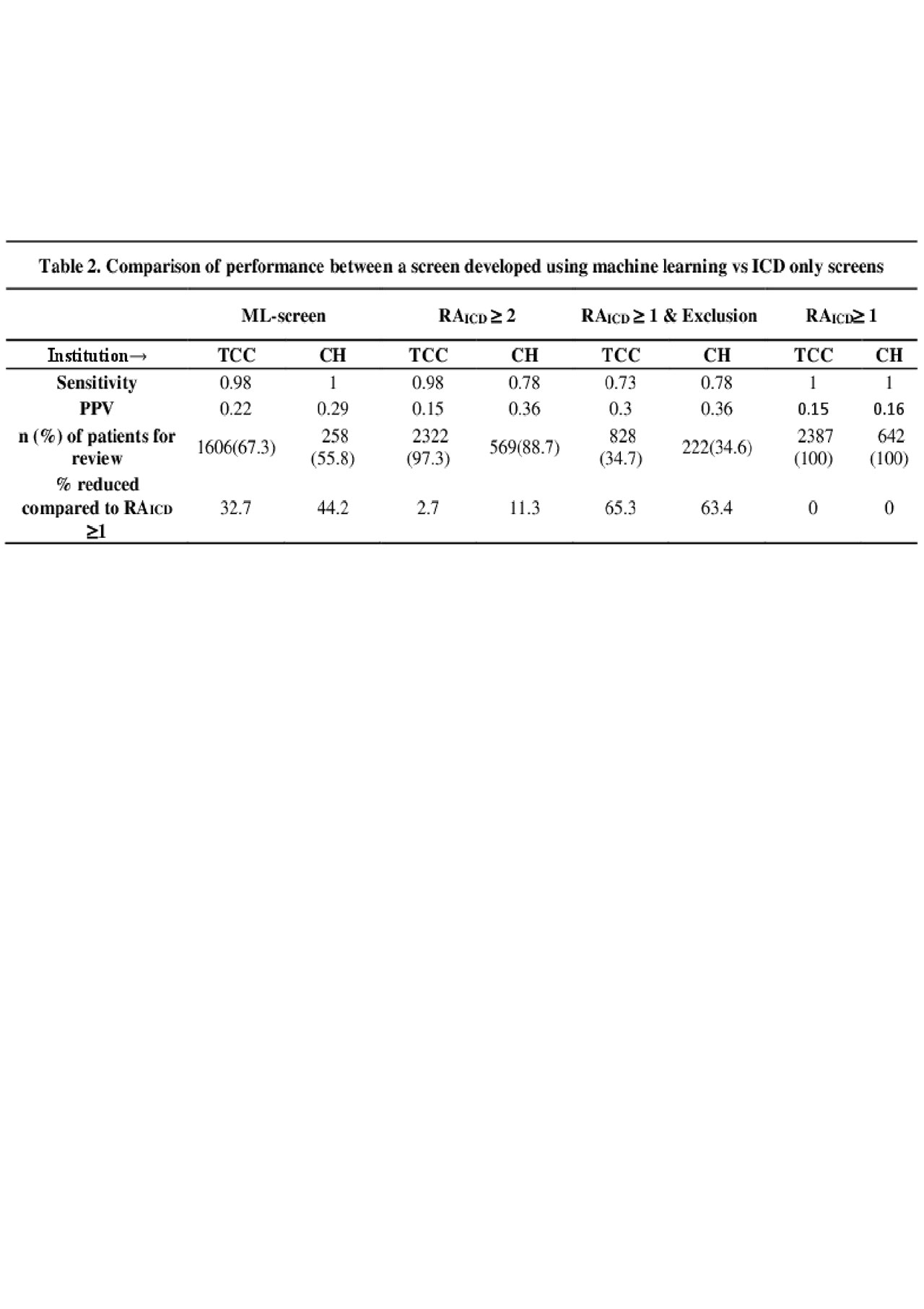
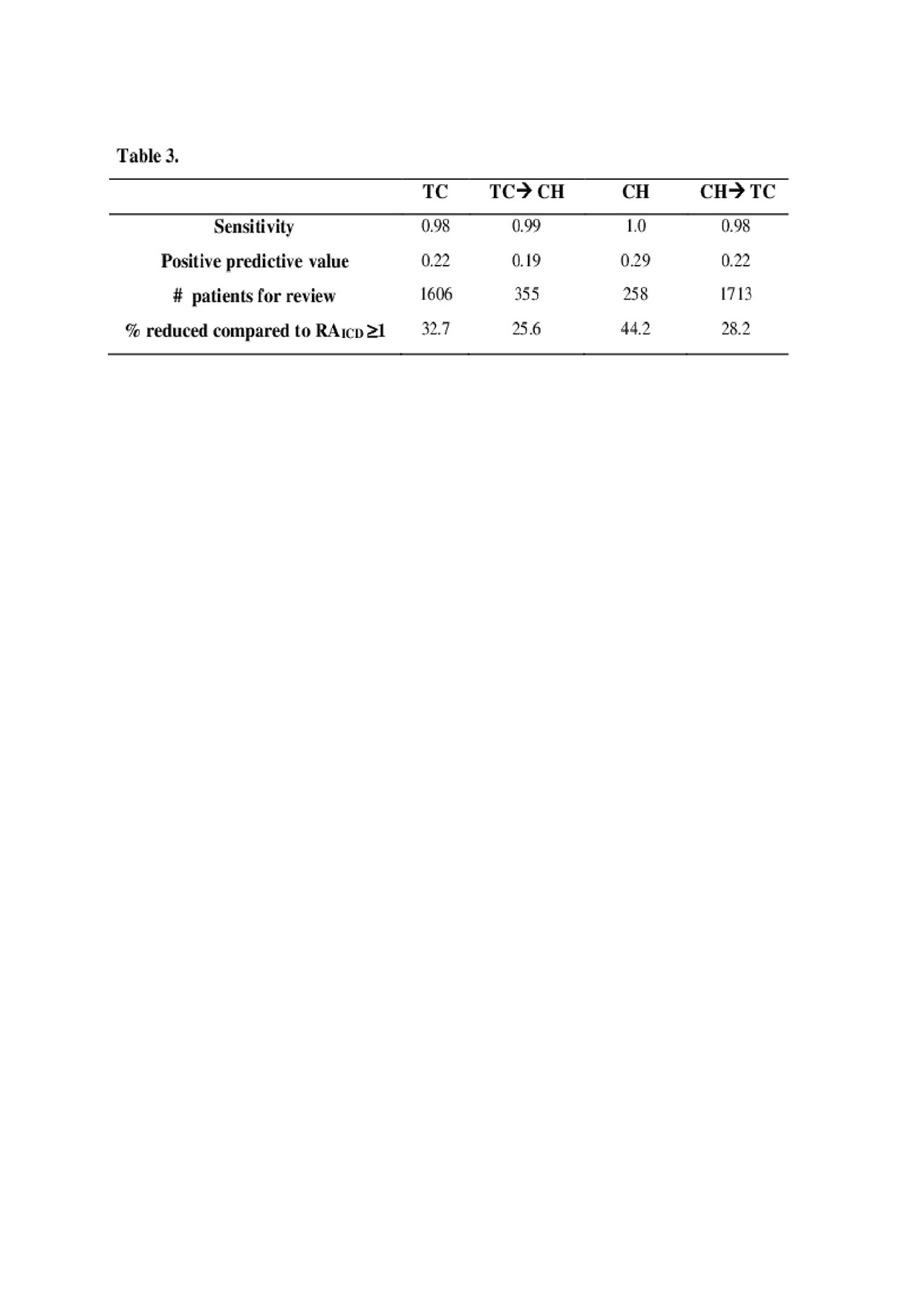
Results: The current method reviewing all charts with RAICD≥1 yielded 346 (14.5%) eligible patients out of 2387 at TCC, and 74 (16.0%) out of 642 at CH. Applying the ML-screen would result in reviewing 33% less patients in TCC and 44% less in CH, compared to RAICD ≥1, without screening out potentially eligible patients (Table 2). In contrast, RAICD ≥2 high sensitivity 0.93-0.98, but did not reduce as many patients for chart review, 2.7-11.3%. The RAICD ≥1+exclusion yielded a larger reduction of patients for review, 63-65%, however excluded approximately 22-27% of eligible patients. The ML-screen had similar performance when trained on one institution and tested on the other (Table 3).
Conclusion: The ML-screen incorporating EHR and NLP data can increase the efficiency of clinical trial recruitment by reducing the number of patients requiring chart review; importantly, this approach did not screen out eligible patients. Moreover, the ML-screen can be trained at one institution and applied at another for multi-center clinical trials.
To cite this abstract in AMA style:
Cai T, Cai F, Dahal K, Hong C, Liao K. Improving the Efficiency of Clinical Trial Recruitment Using Electronic Health Record Data, Natural Language Processing, and Machine Learning [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/improving-the-efficiency-of-clinical-trial-recruitment-using-electronic-health-record-data-natural-language-processing-and-machine-learning/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/improving-the-efficiency-of-clinical-trial-recruitment-using-electronic-health-record-data-natural-language-processing-and-machine-learning/