Session Information
Date: Tuesday, November 14, 2023
Title: (2227–2256) Spondyloarthritis Including Psoriatic Arthritis – Treatment: SpA Poster III
Session Type: Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Approximately 30 to 40% of patients are primary non-responders, and the response rate declines after each successive biological therapy. The primary aim is to determine if cell type-specific transcriptomic data obtained at baseline can predict response to biologics at three months.
Methods: Ethics approval was obtained, and all patients consented to participate. Consecutive patients initiating TNFi or IL-17Ai therapy were assessed using a standardized protocol before initiation of biologic therapy and three months after treatment initiation. Responders were defined as patients with low DAPSA disease activity (≤14). CD8+ T cells were super paramagnetically isolated from PBMCs with Dynabeads (TM) CD8 kit (Cat#11147D). Total RNA was extracted from the CD8+ cells using Lexogen’s Split RNA Kit (Cat#008.48). Libraries were pooled and sequenced on the NovaSeq 6000 for 2 x 150bp reads. STAR (v 2.6) package was used to align raw FASTQ reads and Cufflinks tool (v 2.2.1) was used to quantify the gene expression and then normalized within-sample to transcripts per million (TPM) reads. Differentially expressed genes (DEGs) were detected using limma (v 3.38.3) package. To identify biological pathways that related with drug treatment, the differential expressed genes were ranked by fold change and then to perform Gene Set Enrichment Analysis (GSEA) using MSigDB HALLMARK 2020.
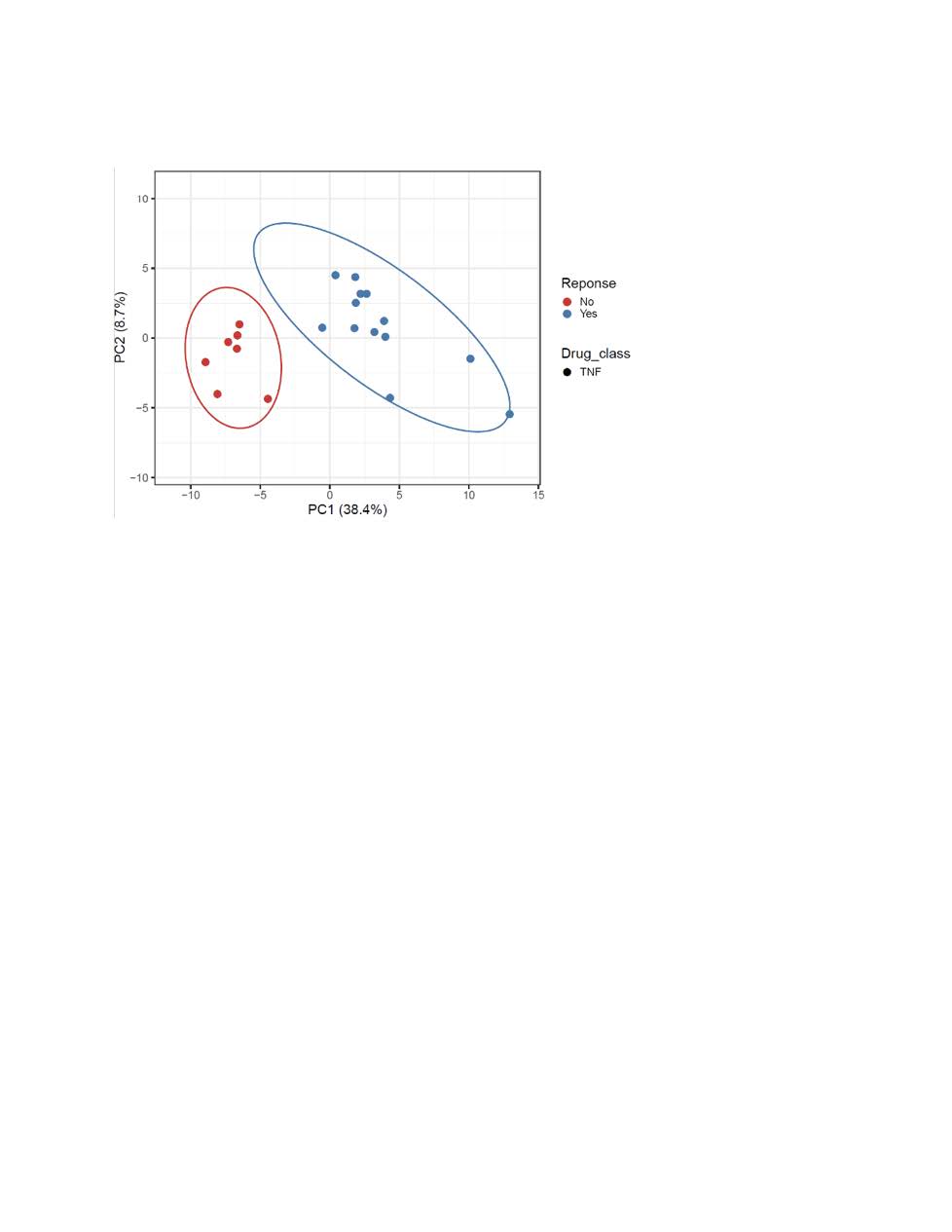
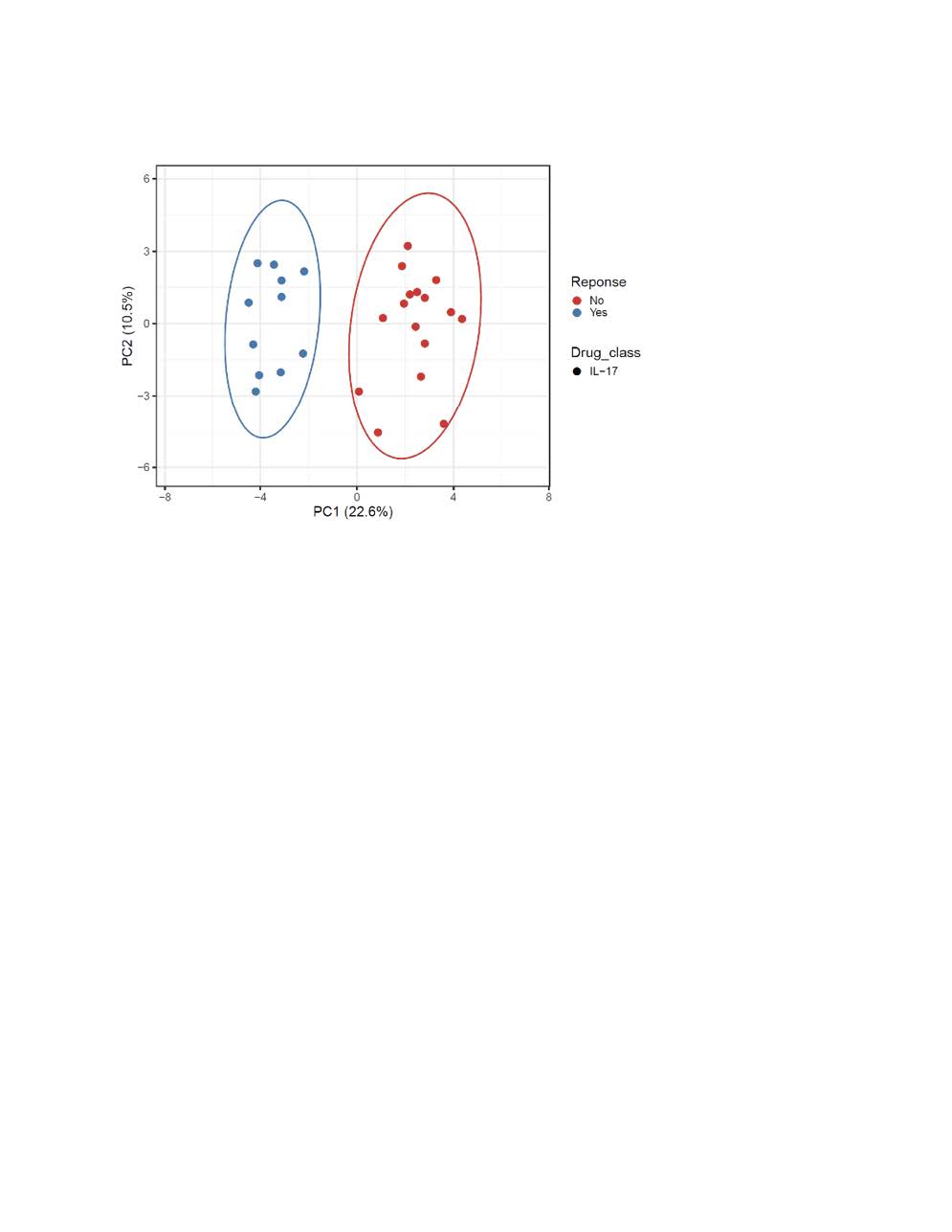
Results: We interrogated 49 PsA patients initiating either TNFi or interleukin-17A inhibitors. Of the 21 TNFi patients the mean age was 56, 14 were females (67%), and 13 (61%) were TNFi responders. For IL-17Ai, there were 28 PsA patients with a mean age of 56, 18 females (64%) and 11 (40%) were responders. PCA analysis discriminated responders vs. non-responder for both TNFi and IL-17Ai. The top five differentially expressed genes for TNFi response were PM20D1, CLIC6, OR1L8, KLHL12, GPALPP1 and for IL-17Ai response were C19orf81, NMI, RPIA, ALG1L, TUBA3E. Using these DEGs, GSEA with the Hallmark gene sets identified MITOTIC_SPINDLE, E2F, G2M as the top down-regulated pathway after IL-17 treatment response, whereas INFLAMMATORY_RESPONSE, INTERFERON_GAMMA_RESPONSE and IL6_JAK_STAT3_SIGNALING remained the top up-regulated pathways before TNFi treatment.
Conclusion: Genomic heterogeneity among PsA patients and cytokine led to differences in expressed genes in different biologic classes. Integration of cell-type-specific DEGs with protein-protein interactions and further comprehensive pathway enrichment analysis is pending.
To cite this abstract in AMA style:
Li Q, O'Rielly D, Jenkins K, Codner D, Gladman D, Dohey A, Jurisica I, Chandran V, Rahman P. Identifying Differentially Expressed Genes to Predict TNF-Alpha and IL-17A Inhibitor Response in Psoriatic Arthritis [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/identifying-differentially-expressed-genes-to-predict-tnf-alpha-and-il-17a-inhibitor-response-in-psoriatic-arthritis/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identifying-differentially-expressed-genes-to-predict-tnf-alpha-and-il-17a-inhibitor-response-in-psoriatic-arthritis/