Session Information
Session Type: Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: Baricitinib is an inhibitor of Jak1 approved for treatment of rheumatoid arthritis, atopic dermatitis, alopecia areata and Covid-19. A phase 2 trial showed success in treating SLE (NCT02708095), but it did not meet its primary endpoint in phase 3 trials(NCT03616964, NCT03616912). To understand the possible dichotomy between these trial results and identify patients with SLE potentially responsive to baricitinib, baseline transcriptomic analysis was carried out in patients enrolled in the phase 2 trial. The goals of this study were to identify the transcriptomic profile of SLE patients who were responsive to baricitinib and the impact of the agent on gene expression abnormalities.
Methods: Baseline whole blood samples from 272 SLE patients from the phase 2 baricitinib trial were utilized for this analysis. These patients had active disease determined by SLEDAI-2K >/=6 and involvement of skin and joints. Patients were randomized to placebo, or one of two doses of baicitinib (2 of 4 mg once daily). Clinical response was determined by perectage of patients resolving skin or joint involvement at 24 weeks using the SLEDAI-2K instrument. RNAseq was performed and analyzed by Gene Set Variation Analysis (GSVA) using 32 informative gene modules and a machine learning (ML) model tested and validated on 3,166 lupus patients from 17 datasets.
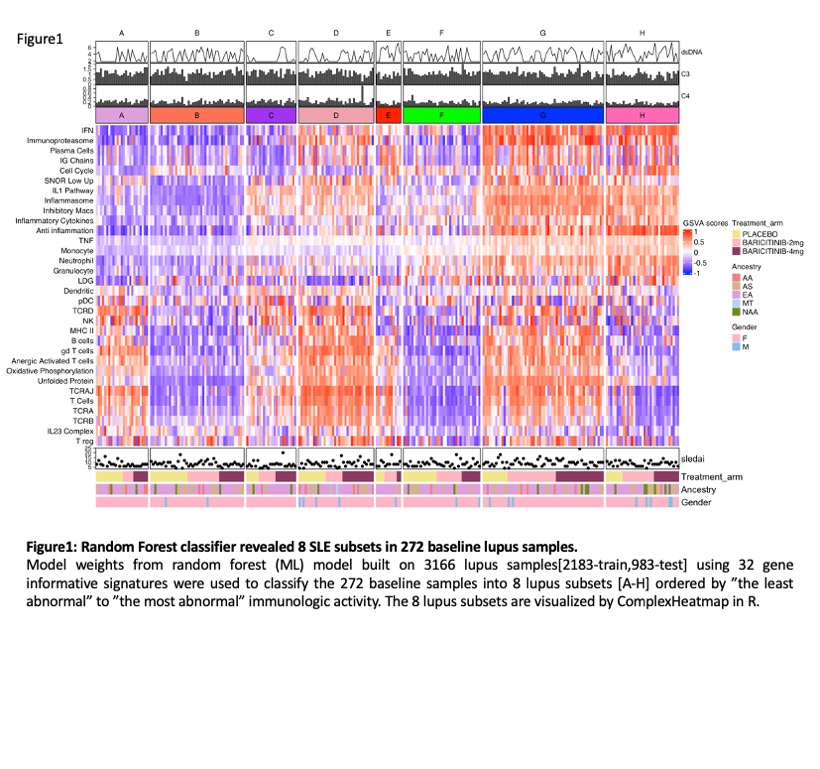
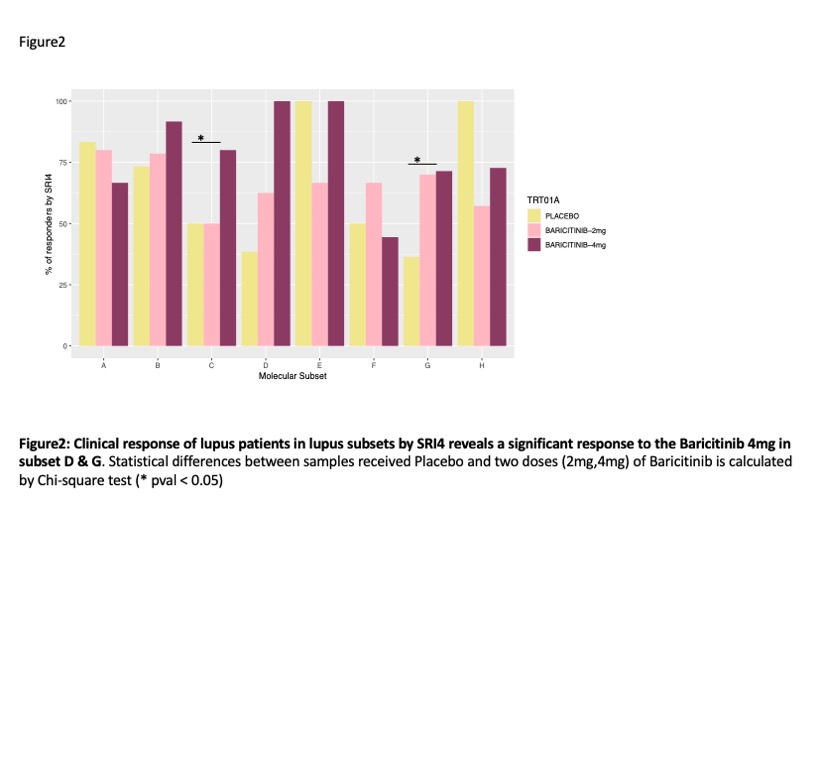
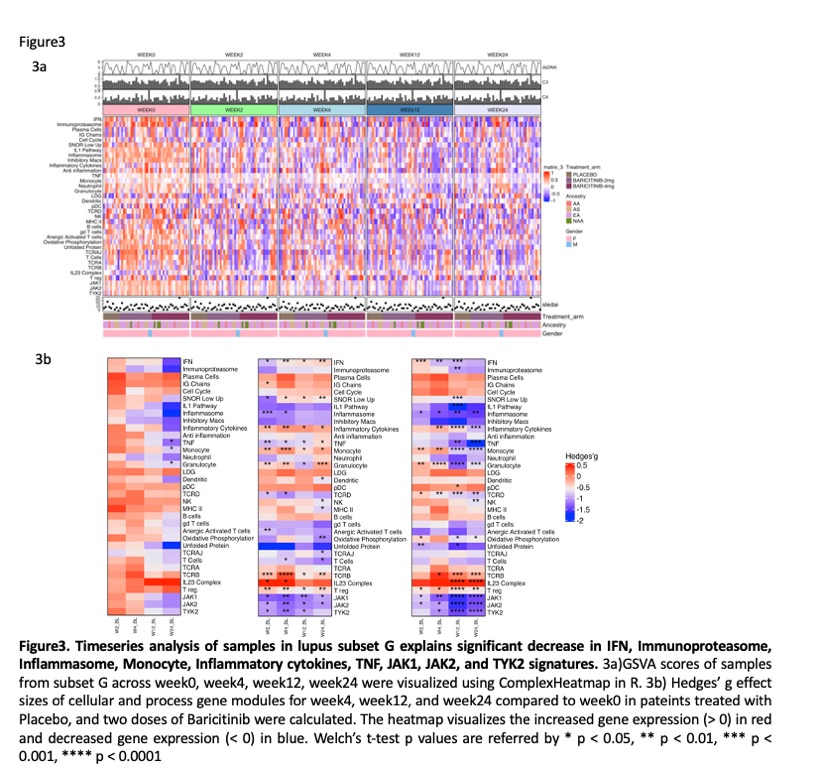
Results: ML analysis of GSVA scores from baseline samples of 272 patients yielded 8 subsets (Figure 1). Subset A had the fewest molecular abnormalities, whereas Subset H had the most disturbances in immune function, including enrichments in the IGS, immunoproteasome, IL-1/ inflammasome pathway, and neutrophil/granulocyte genes and lymphopenia. Clusters B-G had intermediate degrees of abnormal enrichment in specific gene modules. No significant differences in baseline demographic characteristics were noted between the subsets. There were significant but modest differences between subsets in baseline anti-DNA and complement C3 and C4, but no differences in SLEDAI-2K scores. Significant clinical responses to baricitinib were confined to subsets D and G (Figure 2). Effect sizes of responses in these groups were > 30%. Other subsets had higher placebo responses and no additional response to baricitinib. Treatment with baricitinib resulted in significant decreases in the interferon, immunoproteasome, inflammasome, monocyte, granulocye and inflammatory cytokine signatures among others, with improvement observed within 4 weeks of treatment onset. (Figure 3).Notably, gene modules downstream of JAK1, JAK2 and TyK2 were significantly decreased by baricitinib, but not placebo treatment.
Conclusion: Transcriptomic analysis of of baseline samples using GSVA scores from informative gene modules and ML successfully clustered patients into subsets that exhibited differences in response to baricitinib treatment. Treatment with baricitinib altered gene expression profiles in a manner consistent with the known action of the agent. Gene expression based subsetting may be useful to enrich trials for responsive patients and monitor the impact of therapy.
To cite this abstract in AMA style:
Bachali P, Grammer A, Lipsky P. Identification of Subsets of SLE Patients Responsive to Baricitinib by Transcriptomic Analysis at Baseline [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/identification-of-subsets-of-sle-patients-responsive-to-baricitinib-by-transcriptomic-analysis-at-baseline/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-subsets-of-sle-patients-responsive-to-baricitinib-by-transcriptomic-analysis-at-baseline/