Session Information
Date: Tuesday, October 23, 2018
Title: Systemic Lupus Erythematosus – Etiology and Pathogenesis Poster III
Session Type: ACR Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Although many factors have been implicated to be involved in the development of systemic lupus erythematosus (SLE), the pathogenesis of this prototypical autoimmune disease has not yet been fully clarified. Here we are aimed to search for new candidate genes that contribute to SLE disease activity.
Methods: A microarray approach using the Affymetrix Hugene chip was employed for the determination of differentially expressed mRNA profiles between active SLE patients (with SLEDAI score > 8) and normal subjects. The expression of screened key genes was then verified by real-time quantitative polymerase chain reaction (RT-PCR) in peripheral blood mononuclear cells (PBMCs) from 69 SLE patients, 43 normal subjects and 40 patients with other connective tissue diseases. Clinical data of SLE patients were collected to analyze for the relationship with gene expression levels. To identify the cell type involved in specific gene production, monocytes, lymphocytes, eosinophils, neutrophils, T cells and B cells were separated from peripheral blood by flow sorting, and measured for the gene expression by RT-PCR.
Results: Gene chip analysis revealed 750 down-regulated and 597 up-regulated genes in SLE patients, among which RNASE2 (ribonuclease A2), a gene rarely reported previously and highly expressed in SLE group (fold change 6.27, p = 3*10-7), was selected for further study. RT-PCR confirmed that RNASE2 mRNA expression was elevated in PBMCs from SLE patients compared with that from normal subjects as well as patients with primary Sjögren’s syndrome or rheumatoid arthritis (Figure 1). RNASE2 mRNA level was positively correlated with SLEDAI score (r =0.36, p <0.01), BILAG score (r =0.38, p <0.01) and level of 24 hour proteinuria (r =0.40, p <0.01). In addition, the expression level of RNASE2 was related to serum total immunoglobulin G level (r =0.25, p <0.05) and elevated in anti-Sm positive group and anti-dsDNA positive group (both p < 0.01). The cell types mainly involved in RNASE2 production were quite different between normal subjects and SLE patients, which were neutrophils and monocytes respectively.
Conclusion: RNASE2 is highly expressed in monocytes from SLE patients, and correlated with disease activity as well as autoantibody levels. Such insight into SLE pathogenesis may suggest a novel therapeutic target for the disease in the future.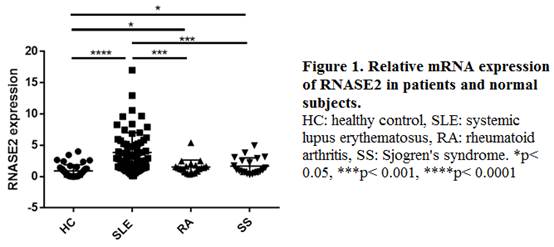
To cite this abstract in AMA style:
Xu Y, Tang X, Feng X. Identification of RNASE2 As a Novel Lupus Candidate Gene [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/identification-of-rnase2-as-a-novel-lupus-candidate-gene/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-rnase2-as-a-novel-lupus-candidate-gene/
