Session Information
Date: Sunday, November 10, 2019
Title: 3S105: Osteoarthritis & Joint Biology – Basic Science (886–891)
Session Type: ACR Abstract Session
Session Time: 4:30PM-6:00PM
Background/Purpose: Erosive osteoarthritis (EOA) of the hand is a characterized radiographically with centrally-located interphalangeal joint erosions, and clinically by its rapid onset, rapid rate of progression, and potential for substantial disability and deformity. Although an association between a single nucleotide polymorphism in the interleukin-1β gene has been demonstrated for EOA in a small case-control study, it remains unclear whether this condition has a genetic basis and there remains a lack of understanding of the underlying pathophysiology. Our purpose was to determine whether EOA clusters in families, which would suggest a genetic etiologic contribution. Secondarily, we performed genomic analyses on one high-risk family with EOA to identify candidate genes that have a significant contribution to the onset and progression of EOA.
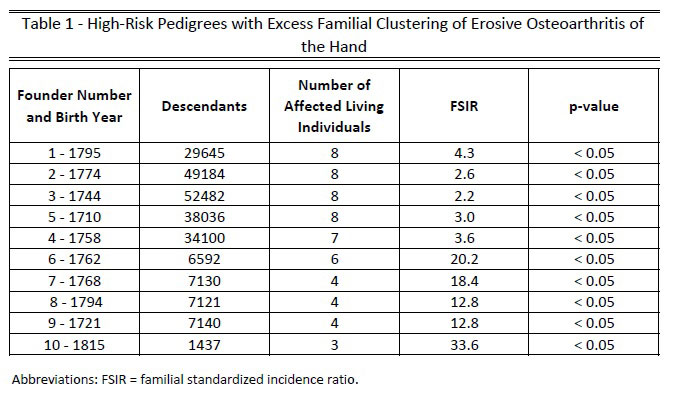
Methods: EOA patients were identified by ICD-10 code (M15.4), then mapped to pedigrees using methods developed by the Utah Population Database (UPDB). The UPDB is a collection of >10 million individuals in multigenerational pedigrees dating back to the late 1700’s which are linked to >30 million medical records. High-risk families with excess clustering of EOA were identified using the Familial Standardized Incidence Ratio (FSIR) threshold of ≥ 2.0. The magnitude of familial risk was calculated using relative risk from Cox regression models to determine the relative risk of EOA in related individuals using a ratio of 1:10 affected individuals to controls. Genomic analyses were performed as previously reported.
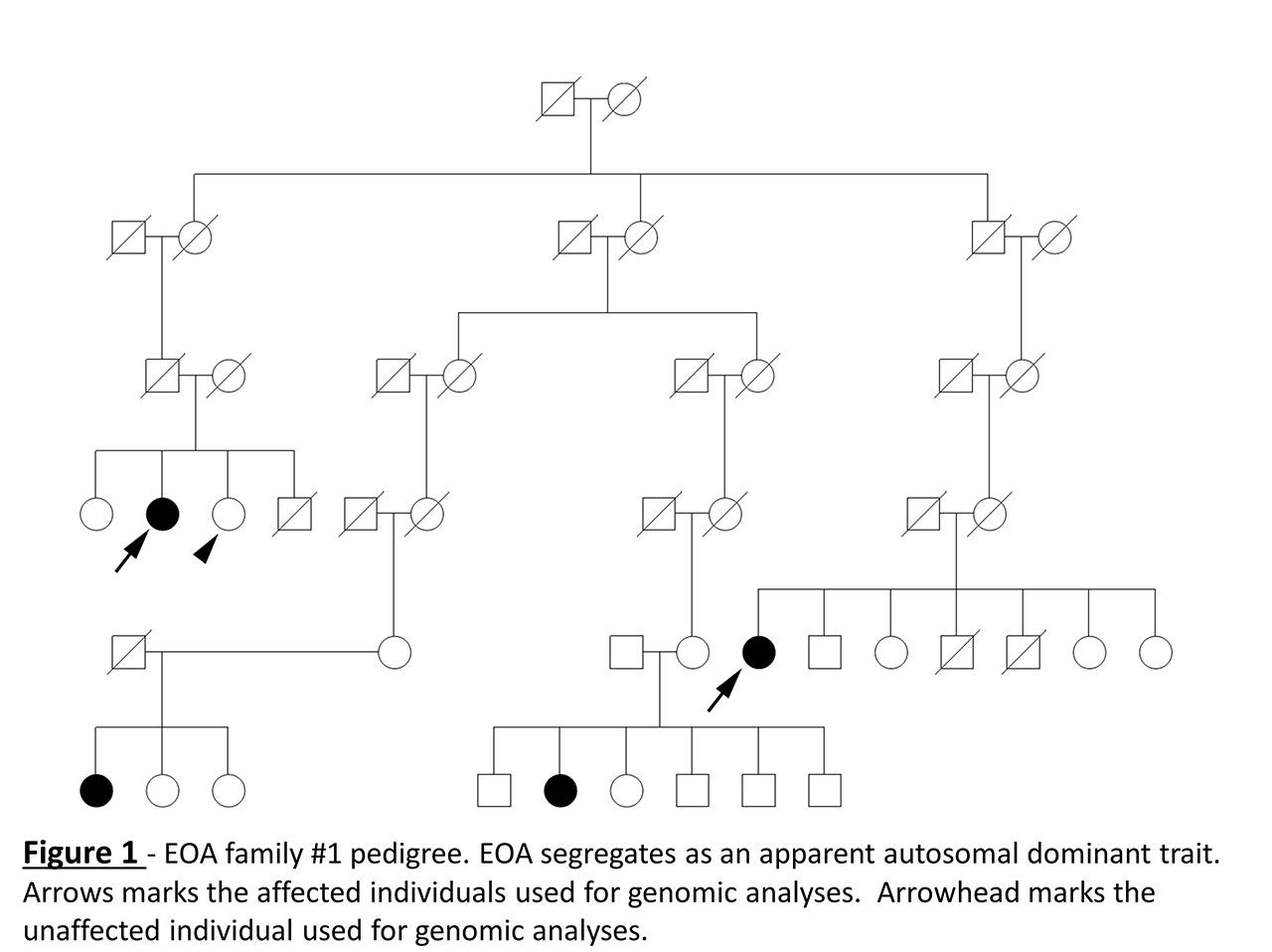
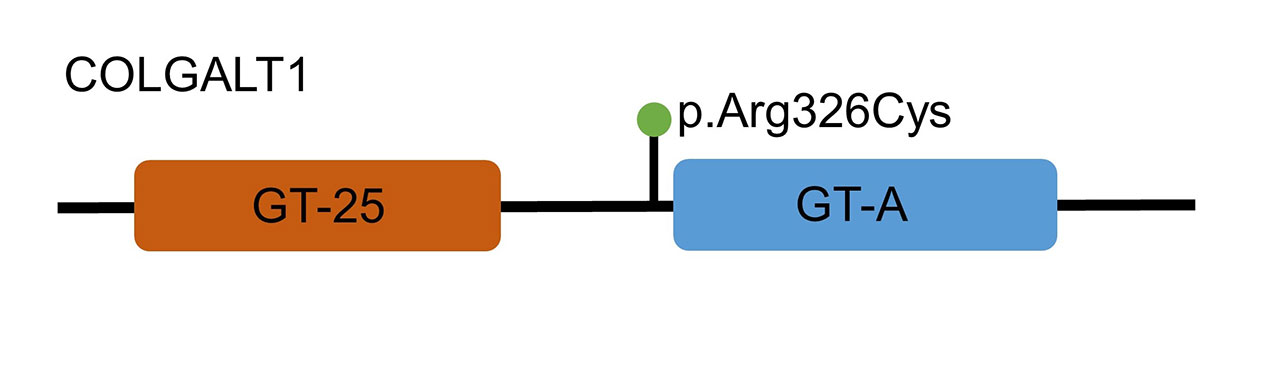
Results: We identified 231 unrelated high-risk pedigrees with an FSIR ≥ 2.0 (top pedigrees illustrated in Table 1). Of the 580 affected individuals within these pedigrees, mean age at diagnosis was 66 ± 11.0 years and 80.1% were female. The relative risk of developing EOA was significantly elevated in first-degree relatives (RR 30.51; 95% confidence interval 3.3 – 282.4; p = 0.0026). Genomic analyses in one EOA family consisting of two distantly related affected individuals and one unaffected sibling (Figure 1) identified a rare coding variant in the procollagen galactosyltransferase COLGALT1 (Arg326Cys, AF – 0.00009) (Figure 2).
Conclusion: Our finding of excess familial clustering of hand EOA patients indicates a significant genetic contribution to the etiology of the disease. Although functional testing is still underway, we have identified a coding variant associated with EOA – namely COLGALT1, a gene involved with glycosylation of hydroxylated lysines in procollagen.6 These results, and those to be gained through whole-exome sequencing of additional identified high-risk pedigrees, may lead to an improved understanding of EOA pathophysiology.
To cite this abstract in AMA style:
Kazmers N, Novak K, Yu Z, Barker T, Abraham T, Romero R, Jurynec M. Identification of Genetic Variants Associated with Erosive Hand Osteoarthritis Using Pedigrees from a State-Wide Population-Based Cohort [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/identification-of-genetic-variants-associated-with-erosive-hand-osteoarthritis-using-pedigrees-from-a-state-wide-population-based-cohort/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-genetic-variants-associated-with-erosive-hand-osteoarthritis-using-pedigrees-from-a-state-wide-population-based-cohort/