Session Information
Session Type: Poster Session (Tuesday)
Session Time: 9:00AM-11:00AM
Background/Purpose: The pathogenesis of osteoporosis, a common disease with high morbidity1, comprises genetic and environmental factors2. Recent studies demonstrated that blood samples are a source of reliable biomarkers which could serve as drug targets in treating of multiple age-associated diseases. The aim of this study was analyze transcriptomes of elderly women with osteoporotic Vertebral Frature, comparing with women with no Vertebral Fracture. Applying bioinformatical tools enabled us to identify such biomarkers for further validation.
Methods: We conducted microarray assays for comparing RNA expression of female vertebral fracture patients and no vertebral fracture controls. Age, bone mineral density (BMD) at lumbar spine, total hip and femoral neck and bone turnover markers were similar between the groups. In statistical analysis of microarrays we applied logistic regression model with age used as a covariate. Bioinformatic analysis involved interactome network analysis in Cytoscape software and enrichment analysis of biological processes regulated by the candidate genes. Special focus was put on identification candidate genes showing also age related changes in expression in bone and muscle tissue according to public available data (GTEX, String database). Expression changes were validated using SYBR green based qPCR-RT (quantitative real-time polymerase chain reaction). We used Pfaff method for relative expression quantification. The geometric mean of 2 control genes (RPL27 and RPL6) was used to compare the gene expression between the groups. Genes with p≤ 0.01 were considered statistically significant in microarray study and p≤ 0.05 in qPCR.
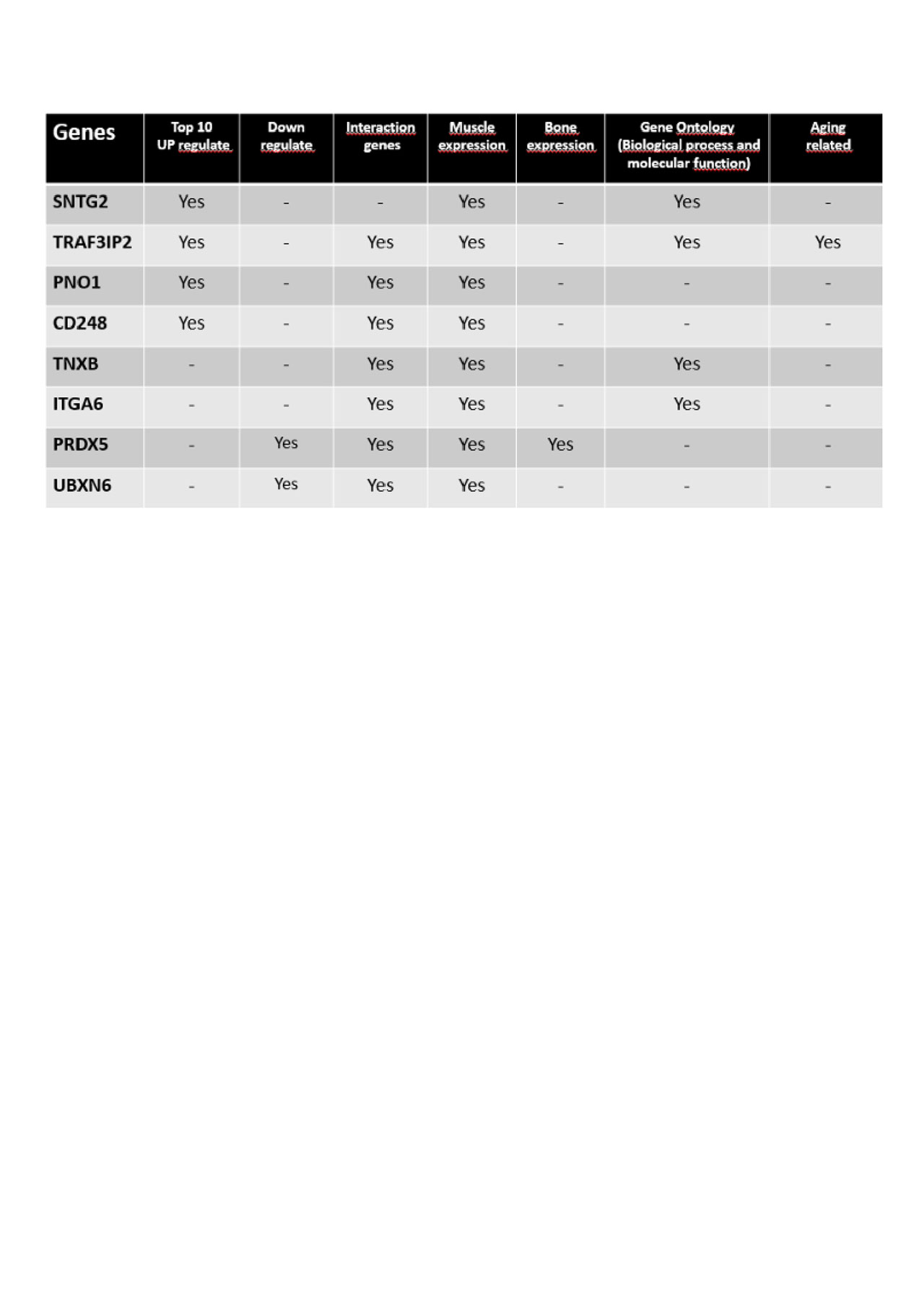
Results: We identified 142 differentially expressed transcripts, 57 up regulated, 85 down regulated in microarray analysis. The transcripts SNTG2, TRAF3IP2, PNO1, CD248, TNXB, ITGA6, PRDX5 and UBXN6 were chosen for validation due to their significance in the analysis of enrichment and genetic interaction. qPCR validation confirmed increased expression in Vertebral Fracture group of TRAF3 Interacting Protein 2 (TRAF3IP2 with fold change = 1.83, SD = 0.66, p = 0.01), Integrin Subunit Alpha 6 (ITGA6 with fold change = 1.62, SD = 0.45, p = 0.007) and Sintrophyn (SNTG2 with fold change = 2.74, SD = 1.68, p = 0.024).
Conclusion: TRAF3IP2 plays a role in various autoimmune and inflammatory diseases and interacts with TNF receptor-associated factor 33. ITGA6 recently was found as upregulated and potentially involved in activity of osteoblasts of rheumatoid arthritis4. SNTGβ2 transcript that encodes a protein belonging to the syntrophin family, highly expressed in the musculoskeletal system5.
Our data support the association of these transcripts with osteoporotic vertebral fracture in elderly women, independently of bone mineral density. These transcripts could be used as biomarkers or therapeutic targets in osteoporotic vertebral fracture in futures studies.
To cite this abstract in AMA style:
Jales Neto L, Wicik Z, Torres G, Takayama L, Lopes N, Pereira a, Pereira R. Identification and Validation of Transcriptional Genes Associated with Osteoporotic Vertebral Fractures by Microarray Study, in Community Elderly Women [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/identification-and-validation-of-transcriptional-genes-associated-with-osteoporotic-vertebral-fractures-by-microarray-study-in-community-elderly-women/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-and-validation-of-transcriptional-genes-associated-with-osteoporotic-vertebral-fractures-by-microarray-study-in-community-elderly-women/