Session Information
Date: Friday, November 6, 2020
Session Type: Abstract Session
Session Time: 5:00PM-5:50PM
Background/Purpose: Delays in diagnosis of psoriatic arthritis (PsA) are common and impair quality of life and function. The study objective was to identify phenotypes, genetic variants, and plasma biomarkers that may predict PsA in patients with psoriasis.
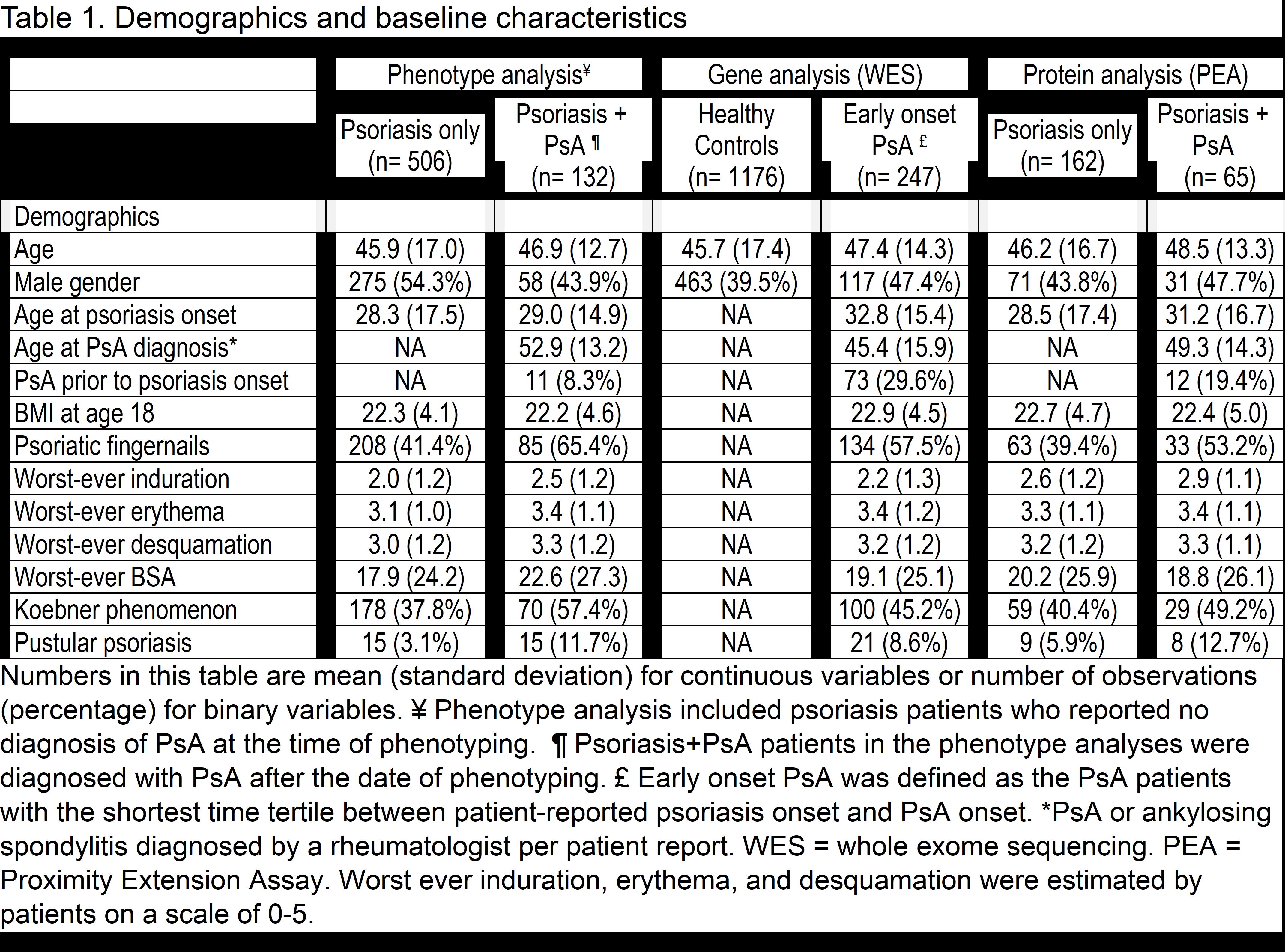
Methods: Clinical data and specimens were collected from 1360 psoriasis patients participating in the Utah Psoriasis Initiative (UPI) between 2002 and 2014. Three concurrent analyses were performed for identifying 1) phenotypes, 2) genes, and 3) plasma protein biomarkers associated with PsA. For the phenotype analysis, proportional hazard Cox regression was used to identify variables associated with an increased risk of developing PsA in the subset of patients with psoriasis phenotyping prior to PsA diagnosis. For the genotype analysis, whole exome sequencing (WES) and primary data analysis were performed by the Regeneron Genetics Center. Variant filtering was completed with the deleterious score BayesDel. Participants with early-onset PsA were compared to healthy controls using an in-house software suite PERCH. Cox regression was also used to assess genotype associations in PsA patients vs. psoriasis patients. For the biomarker analysis, 273 proteins were quantified with Proximity Extension Assay (PEA) and compared in psoriasis subsets with and without PsA.
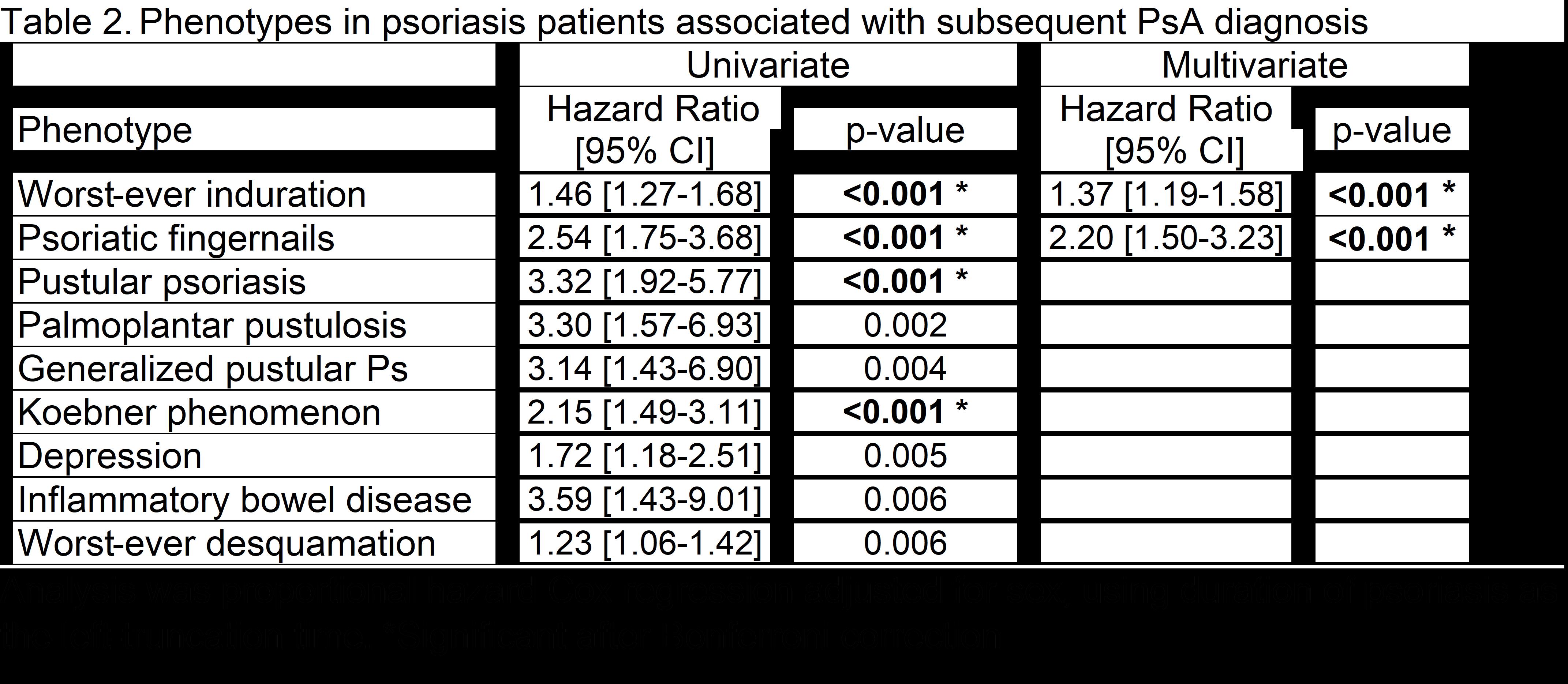
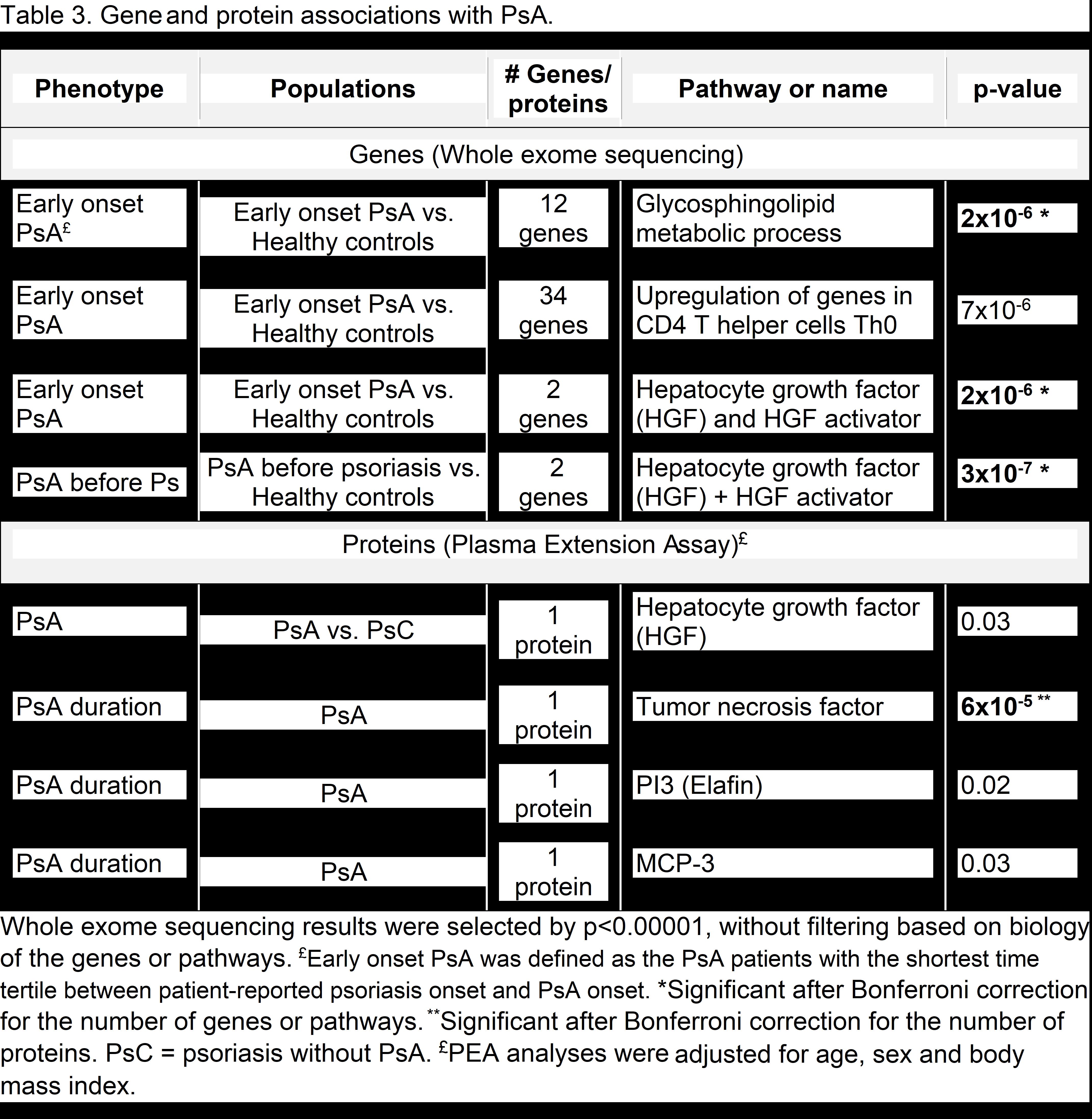
Results: The mean ages of the phenotype, genotype, and biomarker populations ranged from 45.9 to 48.5 and 39.5% to 54.3% were male (Table 1). Phenotypes associated with a subsequent diagnosis of PsA in the multivariate analysis included worst-ever induration and fingernail involvement (Table 2). With WES analysis, early-onset PsA had a significantly higher burden of deleterious or loss-of-function variants in the glycosphingolipid metabolic process pathway and the gene set hepatocyte grow factor (HGF) and HGF activator (HCFAC) than healthy controls (Table 3). The association of HGF+HGFAC was genome-wide significant among the subset of patients who developed PsA before psoriasis. Cox regression in PsA (n=402) vs. psoriasis patients (n=1019) also demonstrated that HGF and HGFAC associated with an increased risk of PsA (p=0.01). With PEA, associations were newly discovered between 1) PsA and HGF and 2) PsA duration and TNF, P13, MCP-3 proteins.
Conclusion: Multiple phenotypes, genes, and proteins associate with PsA. Patient-reported psoriatic nails and worst-ever induration associate with the subsequent development of PsA in psoriasis patients. Genetic analyses demonstrate the newly discovered associations with HGF and HGFAC genes that differentiated between PsA and cutaneous psoriasis. The association between PsA and plasma HGF concentration also suggests that HGF may be a useful protein biomarker for PsA. Additional plasma protein biomarkers for PsA may include TNF, PI3, and MCP-3. After validation and additional discovery of phenotypes, genotypes, and biomarkers, we anticipate using these variables to develop a predictive model for identifying psoriasis patients at high risk for PsA.
To cite this abstract in AMA style:
Walsh J, Belman S, Carroll C, Milliken M, Haaland B, Callis Duffin K, Krueger G, Feng B. Genomic, Phenomic, Proteomic Predictors of Psoriatic Arthritis [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/genomic-phenomic-proteomic-predictors-of-psoriatic-arthritis/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genomic-phenomic-proteomic-predictors-of-psoriatic-arthritis/