Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Childhood-onset systemic lupus erythematosus (cSLE) is a multisystem autoimmune disease with significant morbidity and mortality. Genome-wide association studies have identified more than 100 variants associated with SLE development in adults, but there remains limited data related to genetic variation within cSLE. The aims of this study are to investigate genetic factors of early-onset cSLE, defined as onset of cSLE prior to age 10, and which represent a particularly promising target for variant discovery.
Methods: Employing a case-only design with subgroup analysis (presence of lupus nephritis), we performed Whole Genome Sequencing (WGS) analysis on 37 subjects with early onset cSLE with the hypothesis that rare and/or novel functional variants with large effects in genes associated with SLE contribute to risk of early-onset cSLE diagnosed prior to age 10. We further hypothesized that the polygenic risk score (PRS) for early-onset cSLE patients would be inversely associated with age of onset and presence of nephritis. 153 linkage disequilibrium (LD) independent variants identified in our cohort were compared to previously SLE-reported single-nucleotide polymorphisms (SNPs) and were used to calculate a PRS for our samples. For gene set enrichment analysis, we prioritized rare (MAF ≤ 1%) protein-altering damaging (CADD ≥ 20) and likely pathogenic or pathogenic (ClinVar) variants. A burden score was created based upon these 53 rare pathogenic variants and tested for differences between cases (nephritis versus non-nephritis) using a logistic regression model, including the following covariates: disease activity (SLEDAI), polygenic risk score, sex and age. Gene-based associations were considered significant if FDR< 0.05.
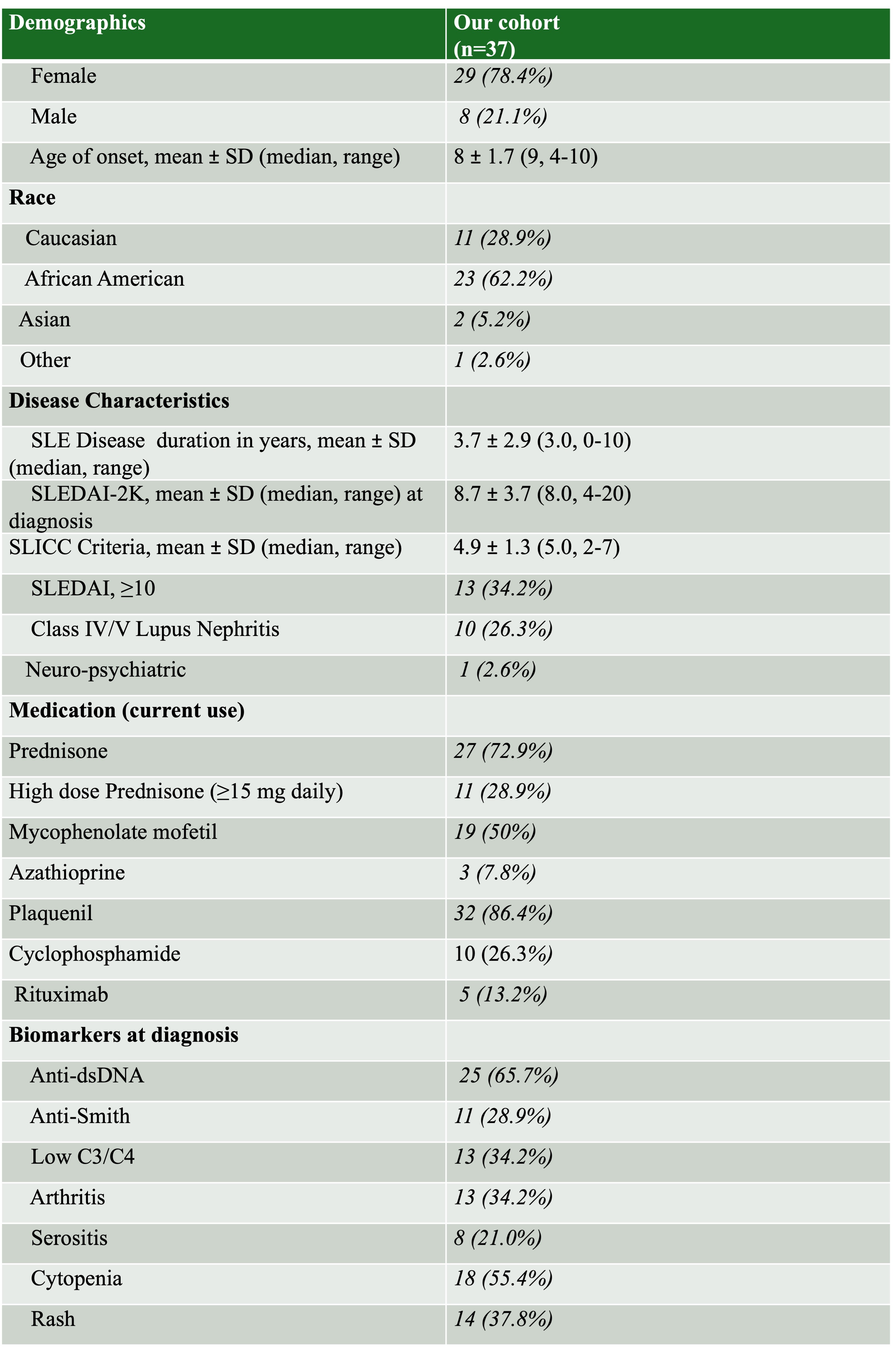
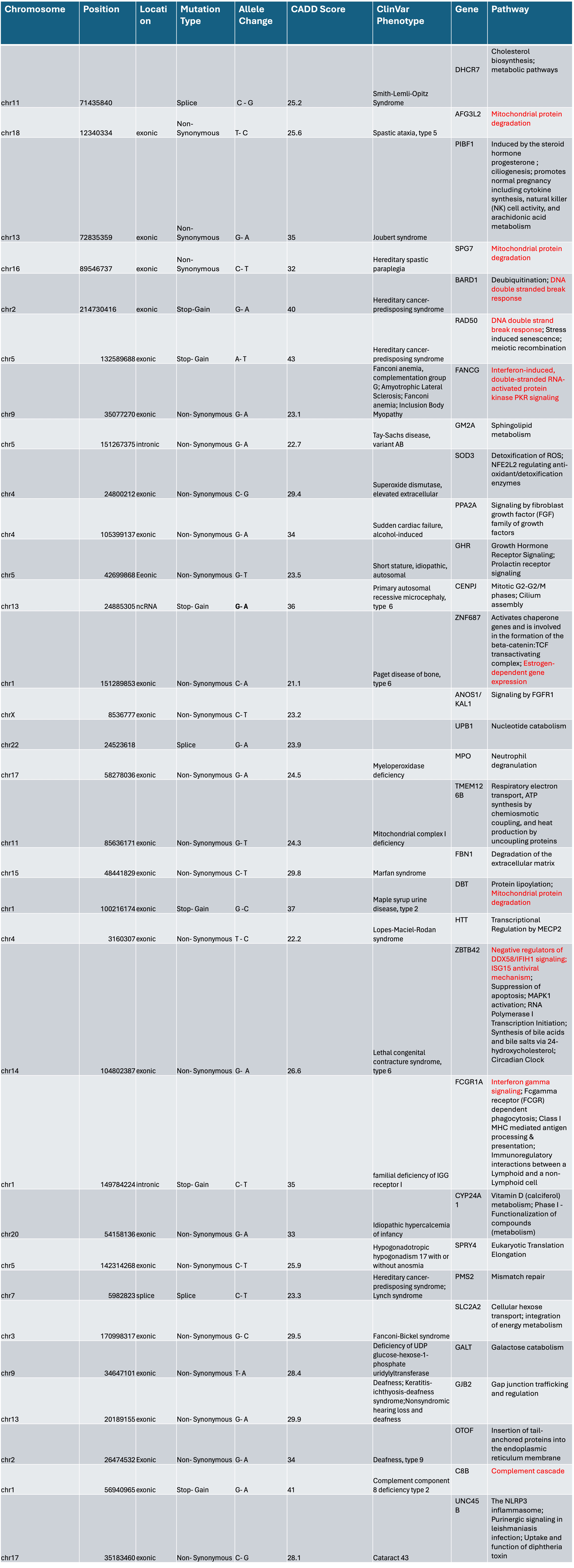
Results: Our cohort analyzed 37 patients, of whom 79% were female. Our cohort had a mean age at diagnosis of 8 ± 1.7 years (Table 1). We identified rare, damaging pathogenic variants in 81% of our patients. Genome wide sequencing identified 868,586 unique variants that overlapped with recently published SLE GWAS data in adults with SLE. PRS calculated for cSLE patients ranged from -0.49 to 0.77 with a median of 0.12. Given our limited statistic power, we did not find a strong inverse relationship between PRS and age of onset (R2=0.08) nor did we find statistically significant findings in gene-set enrichment analysis. However, of the 53 identified rare, damaging, pathogenic variants in our cSLE cohort, these included 3 genes involved in mitochondrial protein degradation (AFG3L2, SPG7, and DBT) and two genes in the interferon pathway (ZBTB42, FCGR1A), well described pathways in lupus pathogenesis (Table 2). We also found an association between age and nephritis, wherein for every increase in year of age in patients with cSLE, the odds for nephritis increased (p=0.0055; Odds Ratio: 2.159, CI: 1.37-4.32). We also found an association between PRS and nephritis, wherein for every unit of increased PRS, the log odds for nephritis increased (p=0.0092).
Conclusion: These results have the potential to enhance our understanding of cSLE and identify important pathways in the pathophysiology of this vulnerable childhood population. Further studies are needed to validate our findings.
To cite this abstract in AMA style:
Nelson M, Murthy S, Maddipatla S, Shenoy S, Ponder L, Kugathasan S, Cutler D, Prahalad S. Genetic Determinants of Childhood Onset Systemic Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/genetic-determinants-of-childhood-onset-systemic-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genetic-determinants-of-childhood-onset-systemic-lupus-erythematosus/