Session Information
Session Type: Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: Patients with incomplete lupus erythematosus (ILE) have features of lupus, but have insufficient criteria for SLE classification. Some ILE patients transition to classified SLE, but the majority remain ILE without major organ involvement. ILE presumably shares genetic associations with SLE, while unique ILE variants or reduced genetic load may influence a more mild disease course. This study aims to identify variants associated with ILE through genome-wide association study (GWAS) and compare genetic load of ILE patients, SLE patients, and unaffected individuals.
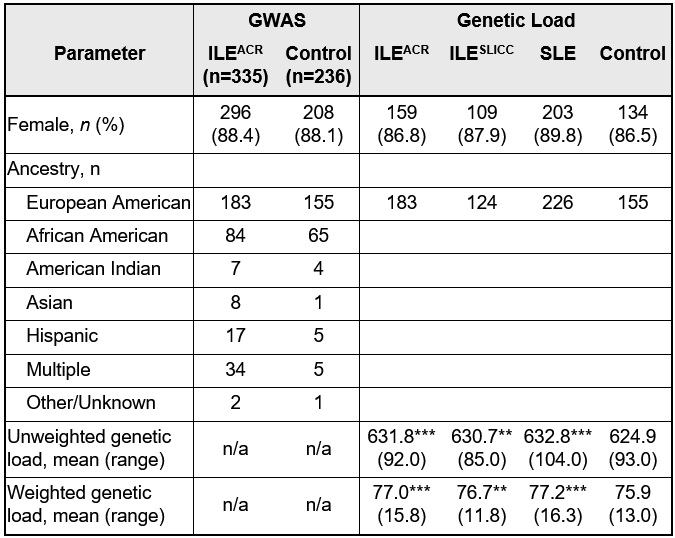
Methods: ILE was defined as 3 ACR-1997 SLE criteria (ILEACR) and SLE as ≥4 ACR-1997 criteria by standardized medical record review. The subset of ILEACR who also did not meet SLE classification by SLICC criteria were designated ILESLICC. Study participants were genotyped on the Infinium Global Screening Array (GSA). After quality control, association testing included 335 ILEACR patients and 236 controls of diverse ancestry without self-reported lupus manifestations (Table). Principal components were included as logistic regression covariates. For genetic load calculations, genotypes were pre-phased and imputed against 1000 Genomes Project haplotypes. Risk alleles that showed association with European SLE in Langefeld, et al (Nat Comm, 2017) (pFDR< 0.05) were identified in unrelated European American (EA) patients with SLE, ILEACR, ILESLICC, and controls (Table). Genetic loads were computed for each person as the number of risk alleles (unweighted) and allele sum with each allele weighted by its beta coefficient (weighted).
Results: Through GWAS in the ancestrally diverse population of ILEACR and controls, 12 variants exceeded the suggestive level of significance (p < 1 × 10−5). Eight were not previously reported in SLE (DPP10-DDX18, LRP1B, HMHB1-YIPF5, PHF14-THSD7A, ABCA1, DMRTA1-LINC01239, LOC105369443-CNTN5, and MIR3649-ADIPOR2), involving genes associated with phospholipid metabolism, asthma, Behçet disease, and cancers. No variants reached genome-wide significance (p < 5 × 10−8) in this small study. EA ILEACR, ILESLICC, and SLE patients had significantly greater unweighted and weighted genetic load than controls (p < 0.001) (p < 0.001) and (p < 0.01) (Table), respectively. There was no significant difference between mean genetic load (weighted or unweighted) in ILE and SLE, and weighted genetic load did not significantly differentiate between ILEACR and SLE in receiver operator characteristics analysis (area under the curve = 0.52). Further, those with a weighted genetic load of 81.0 (+/-2.0) or higher had greater odds of being SLE or ILE vs. healthy control (odds ratios >5.4-6.3), compared to individuals in the lowest weighted genetic load decile (Figure 1-C). However, weighted genetic load did not significantly affect odds of having SLE vs. ILEACR (Figure 1D).
Conclusion: ILE and SLE patients have similar genetic loads of SLE risk loci, suggesting a similar genetic predisposition between these conditions. Suggestive associations for novel loci are identified in ILE patients which require replication and validation. Phenotypic differences may be influenced by novel ILE variants and gene-environment interaction.
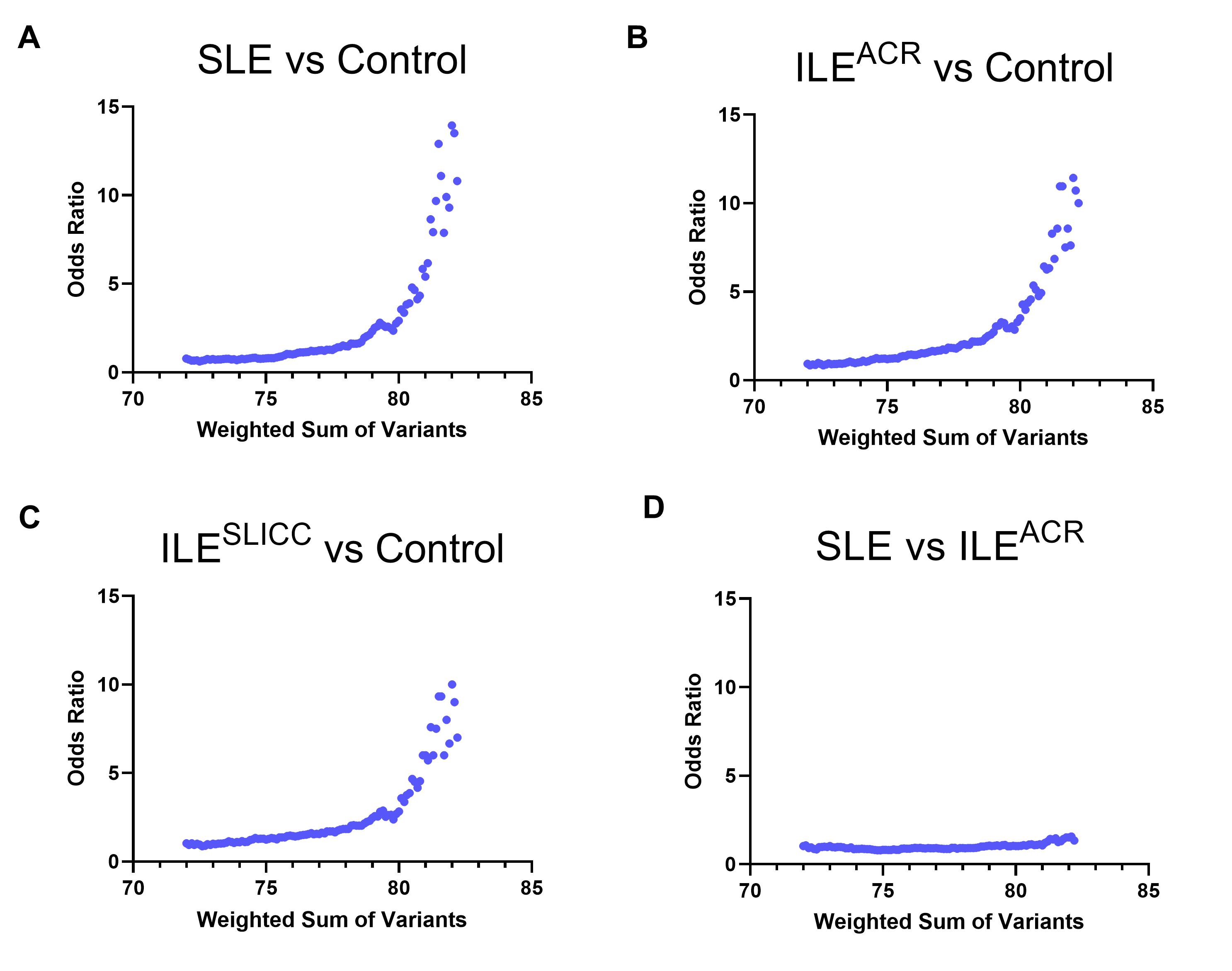 Figure 1. Odds ratio across genetic load range. The odds ratio represents the odds that an individual with a given genetic load (+/-2.0) will have SLE vs control (A), ILE(ACR) vs control (B), ILE(SLICC) vs control (C), or SLE vs ILE(ACR) (D), compared to a baseline comparison group of individuals in the lowest decile for weighted genetic load.
Figure 1. Odds ratio across genetic load range. The odds ratio represents the odds that an individual with a given genetic load (+/-2.0) will have SLE vs control (A), ILE(ACR) vs control (B), ILE(SLICC) vs control (C), or SLE vs ILE(ACR) (D), compared to a baseline comparison group of individuals in the lowest decile for weighted genetic load.
To cite this abstract in AMA style:
Slief M, Levin J, Macwana S, DeJager W, Bourn R, Nath S, Munroe M, Aberle T, Gaffney P, Merrill J, James J, Guthridge J. Genetic Associations and Polygenic Risk Assessment in Incomplete Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/genetic-associations-and-polygenic-risk-assessment-in-incomplete-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genetic-associations-and-polygenic-risk-assessment-in-incomplete-lupus-erythematosus/