Session Information
Session Type: Abstract Submissions (ACR)
Background/Purpose: Psoriasis vulgaris (PsV) risk is strongly associated with genetic variation within the major histocompatibility complex (MHC) region, although its fine genetic architecture has not been elucidated.
Methods: To fully characterize and fine-map the MHC associations of PsV, we conducted a large-scale fine-mapping study of PsV risk in the MHC region in 9,247 PsV cases and 13,589 controls of European descent. We also evaluated risk of two major clinical subtypes of PsV, psoriatic arthritis (PsA; n = 3,038) and purely cutaneous psoriasis (PsC, defined as 10 years of psoriasis without developing PsA; n = 3,098). We imputed class I and II HLA gene variants by applying SNP2HLA software to the SNP genotype data. In addition, we newly constructed an imputation reference panel of sequence variants for MICA, an HLA-like gene within the MHC region that has been implicated for PsA risk. We applied MICA variant imputation to the SNP genotype data and evaluated their risk as well.
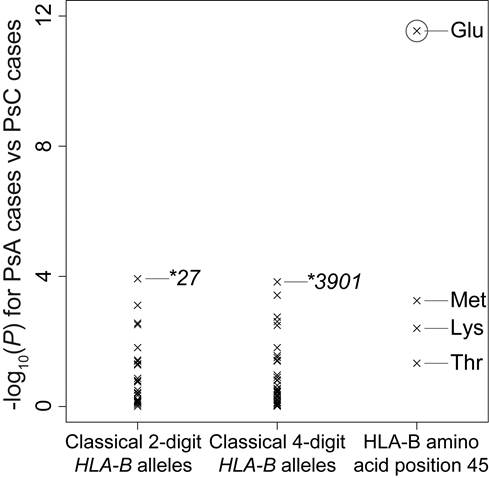
Results: As previously described, we observed that HLA-C*06:02 demonstrated the most significant impact on overall PsV risk (odds ratio [OR] = 3.38, 95% confidence interval [95%CI]: 3.18-3.60, P = 1.7×10-364). Stepwise conditional analysis revealed multiple independent risk variants of both class I and class II HLA genes for PsV susceptibility independent of HLA-C*06:02 (HLA-C*12:03, HLA-B amino acid positions 67 and 9, HLA-A amino acid position 95, and HLA-DQa1 amino acid position 53; P < 5.0x10-8), but no apparent independent risk conferred by MICA. Strikingly, we found that risk heterogeneity between PsA and PsC may be driven by one amino acid position at HLA-B (Glu at HLA-B amino acid position 45; OR = 1.46, 95%CI: 1.31-1.62, P = 2.9×10-12), which demonstrated much more significant association signals compared to classical HLA-B alleles including HLA-B*27 and HLA-B*39:01 (P > 1.0×10-4).
Conclusion: These results indicate that multiple class I and II HLA genes (HLA-C, HLA-B, HLA-A, and HLA-DQA1) contribute to development of PsV, and suggest that different genetic factors, most evident at HLA-B, underlie for the differential risk of specific PsV sub-phenotypes. Our study illustrates the value of high-resolution HLA and MICA imputation for fine-mapping causal variants in the MHC.
Disclosure:
Y. Okada,
None;
B. Han,
None;
L. C. Tsoi,
None;
P. E. Stuart,
None;
E. Ellinghaus,
None;
T. Tejasvi,
None;
V. Chandran,
None;
F. Pellett,
None;
R. Pollock,
None;
A. M. Bowcock,
None;
G. G. Krueger,
None;
M. Weichenthal,
None;
J. J. Voorhees,
None;
P. Rahman,
None;
P. K. Gregersen,
None;
A. Franke,
None;
R. P. Nair,
None;
G. R. Abecasis,
None;
D. D. Gladman,
Abbvie, Amgen, Celgene, Janssen, Pfizer, UCB,
2,
Abbvie, Amgen, BMS, Celgene, Eli Lilly, Janssen, Novartis, Pfizer, UCB,
5;
J. T. Elder,
None;
P. I. de Bakker,
None;
S. Raychaudhuri,
None.
« Back to 2014 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/fine-mapping-major-histocompatibility-complex-associations-identified-contribution-of-multiple-class-i-and-ii-hla-genes-on-risk-of-psoriasis-and-its-clinical-subtypes/