Session Information
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Many genetic and environmental risk factors for SLE are now known. We have developed SLE risk prediction models, incorporating known genetic, lifestyle, and environmental risk factors in the Nurses’ Health Study cohorts (NHS), with area under the receiver operating characteristic (AUC) 0.76 (95% CI 0.72-0.81) for SLE among women. (Cui J, Seminars Art Rheum 2023). Here, we aimed to externally validate and extend these models in a separate population.
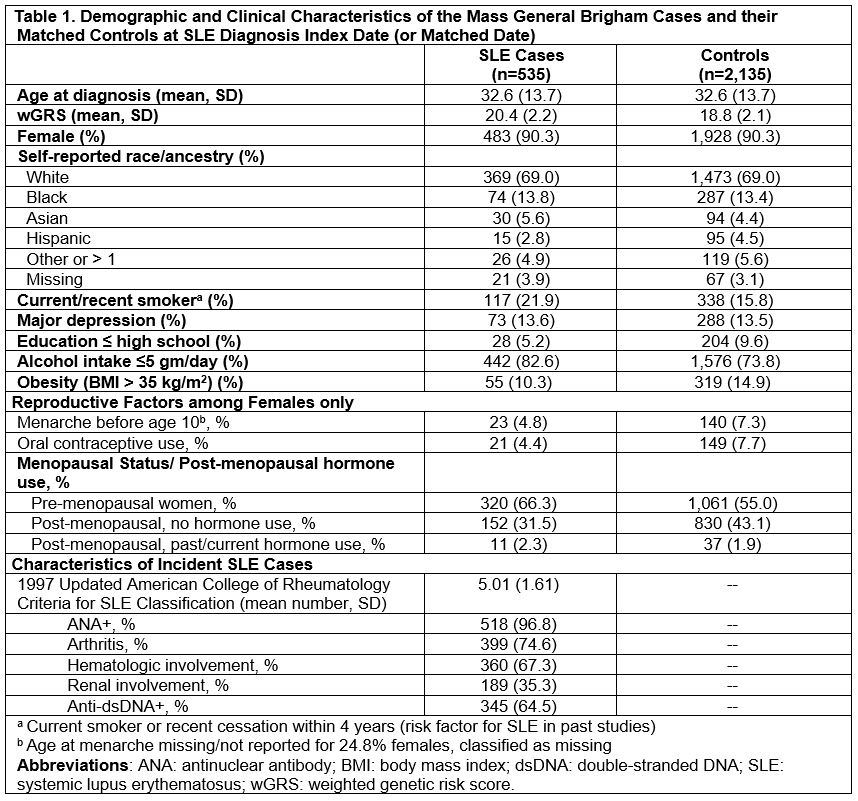
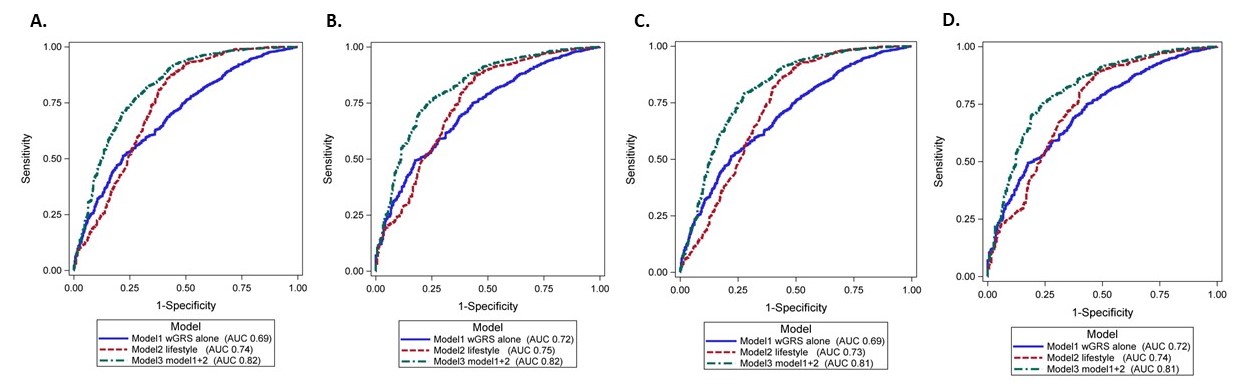
Methods: We performed a nested case-control study in the Mass General Brigham (MGB) Biobank, containing an enrollment health assessment survey, genome-wide genotyping, and linked electronic medical record (EMR) data on 149,680 consented patients. Diagnosed SLE cases met Updated 1997 ACR and/or 2019 EULAR/ACR criteria. SLE patients with genotyping were selected and matched by age (± 4 years), sex, and race/ethnicity to patients without SLE with similar data. As in our prior NHS models, we collected data on risk factors (from prior to SLE diagnosis or matched index date), including current/recent smoking, low/no alcohol use, highest educational level, body mass index, major depression, among females, age at menarche, oral contraceptive use, and menopausal status/postmenopausal hormone use. Our SLE weighted genetic risk score (wGRS) used 86 known single nucleotide polymorphisms (SNPs) and 10 classical HLA alleles. We compared 3 multivariable risk prediction models, for all SLE and for dsDNA+ SLE, and for females only and then including males (without female reproductive factors): Model 1. SLE wGRS; Model 2. current/recent smoking, low/no alcohol use, obesity, depression, race/ethnicity, education status and, among females, early menarche, oral contraceptive use, and menopause/postmenopausal hormones; Model 3. Models 1+2. All models additionally adjusted for matching factors.
Results: We matched 535 patients with SLE (90% female) to 2,135 (90% female) patients without. Index date characteristics are shown in Table 1. Models 1-3 for all SLE among women yielded AUCs 0.69 (95%CI 0.67-0.72), 0.74 (95%CI 0.72-0.77), 0.82 (95% CI 0.79-0.84). Model 3 based on genetics, lifestyle and environmental factors had best discrimination for both SLE and ds+DNA SLE in females only and for both sexes (AUC 0.81 [95% CI 0.78-0.83]).
Conclusion: Our model including SLE wGRS, lifestyle, and environmental risk factors accurately classified future SLE risk with AUC of 0.81-0.82 in this external population. As genetic data are increasingly available and risk factors easily assessed, this or similar SLE prediction model could be feasibly employed for screening at-risk populations for prevention interventions.
To cite this abstract in AMA style:
Koopman J, Cui J, Guan H, Stevens E, Oakes E, Costenbader K. External Validation and Extension of Population-Based Systemic Lupus Erythematosus Risk Prediction Models Using Genetics, and Lifestyle and Environmental Factors [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/external-validation-and-extension-of-population-based-systemic-lupus-erythematosus-risk-prediction-models-using-genetics-and-lifestyle-and-environmental-factors/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/external-validation-and-extension-of-population-based-systemic-lupus-erythematosus-risk-prediction-models-using-genetics-and-lifestyle-and-environmental-factors/