Session Information
Session Type: Poster Session (Monday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Systemic lupus erythematosus (SLE) is an autoimmune disease with heterogeneous clinical manifestations. Epigenetic changes, including DNA methylation, have been implicated in SLE; specifically, differences in methylation have been associated with auto-antibody and lupus nephritis status. A prior study showed that closer residential proximity to highways was associated with hypomethylation of 3 CpG sites in the UBE2U gene in SLE patients.
Methods: In this study, we examine the association of particular matter 2.5 (PM2.5) levels on the day of blood draw with DNA methylation in a cohort of SLE patients from the California Lupus Epidemiology Study (CLUES). All subjects satisfy the ACR criteria for SLE. PM2.5 levels were estimated by Sonoma Technology from air pollution concentrations based on geocoded residential locations. DNA methylation was measured using the Illumina HumanMethylationEPIC BeadChip for 271 unique subjects of White, Hispanic, African American, and Asian ethnicities. Noob background subtraction with dye-bias correction as well as quantile normalization were conducted in minfi. CpG sites with high detection p-values, cross-reactive probes, and CpG sites potentially measuring SNPs were removed for a total of 748,793 CpG sites for analysis. We first conducted an analysis for differentially methylated position (DMPs) using limma linear models with empirical Bayes variance shrinkage, adjusting for age, sex, current smoking status, current medication use, estimated cell-type proportions from ReFACTor, and genetic ancestry. We then conducted an analysis of differentially methylated regions (DMRs) using bumphunter. Given the previous findings in the UBE2U gene, we also conducted a candidate DMP analysis for the 21 CpG sites on the EPIC chip which map to UBE2U.
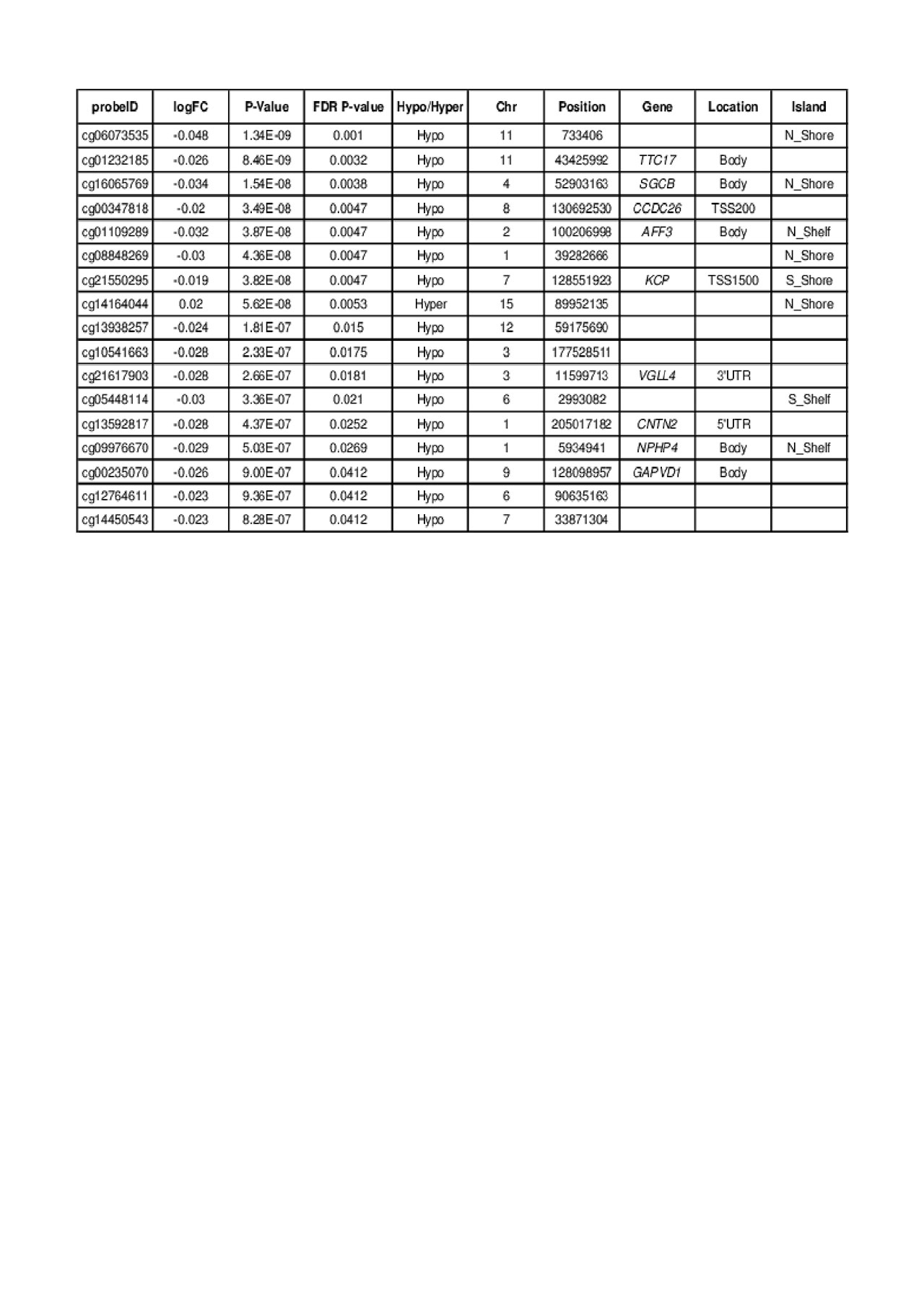
Results: Concentrations of PM2.5 fell between 1.8 and 25.9 μg/m3 (SD = 3.3) with all subjects’ exposure classified as either “Good” or “Moderate” according to the Air Quality Index (AQI). Preliminary results showed 17 DMPs (after FDR correction) associated with PM2.5 exposure, 16 of which were hypomethylated. These CpG sites mapped to genes including TTC17, SGCB, CCDC26, AFF3, KCP, VGLL4, CNTN2, NPHP4, and GAPVD1. One region located 900bp upstream of KLRC3 was significantly differentially methylated after FDR correction. No CpG sites in UBE2U reached statistical significance.
Conclusion: These results suggest for the first time that there is differential methylation with increased PM2.5 exposure in SLE patients.
To cite this abstract in AMA style:
Solomon O, Lanata C, Dall'Era M, Yazdany J, Katz P, Trupin L, Taylor K, Nititham J, Rhead B, Criswell L, Barcellos L. DNA Methylation Changes Are Associated with Particulate Matter 2.5 Exposure in SLE Patients [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/dna-methylation-changes-are-associated-with-particulate-matter-2-5-exposure-in-sle-patients/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/dna-methylation-changes-are-associated-with-particulate-matter-2-5-exposure-in-sle-patients/