Session Information
Session Type: Poster Session (Tuesday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Systemic Lupus Erythematosus (SLE) is a potentially life-threatening autoimmune disease with no cure. The onset of SLE is thought to be the result of environmental events in a genetically susceptible host, yet the underlying genetic architecture of the disease is not fully understood. Pediatric SLE (pSLE) patients have early disease onset and incur more organ damage than adults, and thus may have a larger genetic burden. Confirmed genetic associations with common variants have been demonstrated in many SLE genome wide association studies (GWAS). Examining the polygenic disease score allows us to capture both the number of common SLE risk variants and the magnitude of influence of each genetic variant on SLE predisposition. While a few studies have used a polygenic risk score to explore genetic burden differences in adult SLE patients, this measure has not been reported in pediatric onset SLE.
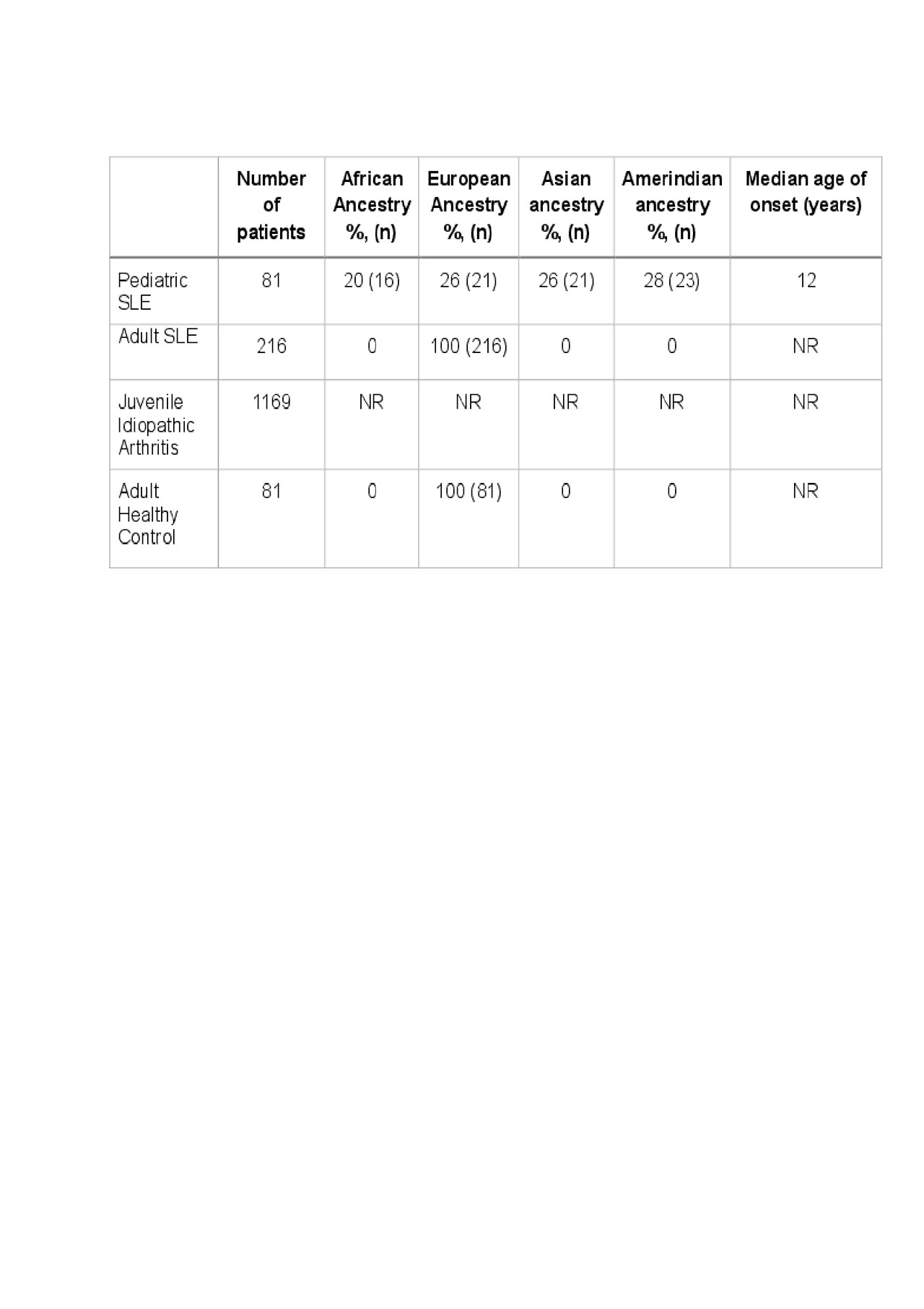
Methods: All SLE patients included fulfilled at least four of the 11 American College of Rheumatology classification criteria for SLE; pSLE subjects met criteria prior to age 18. Samples collected from 81 SLE patients were sequenced via whole genome sequencing using Illumina HiSeq X Ten. Baseline demographics are reported in Table 1. GWAS data from 1169 juvenile idiopathic arthritis patient genotypes were used as pediatric controls, and publicly available GWAS data from adult SLE women and healthy controls were used as a disease specific comparison group. 238 SLE risk single nucleotide polymorphisms (SNPs) tagging independent previously published and established SLE genetic susceptibility loci were assessed in the study. The risk variants were pruned for SNPs in linkage disequilibrium with PLINK 1.9 software using an r2 value of 0.2, resulting in 183 SLE risk loci shared between pSLE and JIA controls. For adults, a limited number of 63 risk loci were shared and used to compare pediatric and adult risk. A polygenic risk score was calculated for each subject based on the sum of the product of the natural logarithm of the odds ratio for each association and the number of alleles present in each individual at each risk locus. Cumulative scores were calculated for all subjects. The case and control means and distributions of polygenic risk score were evaluated with the Wilcoxon rank-sum test.
Results: Pediatric-onset SLE patients have a high burden of SLE common risk SNPs, with mean polygenic risk score of 35.9. Comparing to other children with autoimmune diseases, pediatric onset SLE patients also have a significantly higher polygenic risk score than children with juvenile idiopathic arthritis (35.9 vs. 29.9, p value 2.2 x 10-16). PSLE patients have a similar mean polygenic risk score (11.9) as adult SLE (11.9, p value NS) and higher than healthy adult subjects (11.4, p value 0.002)
Conclusion: Patients with childhood onset SLE have an increased burden of common risk SNPs for SLE when compared to children with non-SLE autoimmune disease. SLE Both childhood and adult-onset SLE have a higher polygenic risk score than adult healthy controls subjects. Childhood onset SLE patients have a polygenic risk score similar to adult onset SLE patient. This may be due to limited number of shared SNPs available for assessment or limited sample size for comparison.
To cite this abstract in AMA style:
Lewandowski L, Ombrello M, Aksentijevich I, Deng Z, Hiraki L, Silverman E, Scott C, Barrera-Vargas A, Hasni S, Kaplan M. Determining a Polygenic Risk Score in Pediatric Systemic Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/determining-a-polygenic-risk-score-in-pediatric-systemic-lupus-erythematosus/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/determining-a-polygenic-risk-score-in-pediatric-systemic-lupus-erythematosus/