Session Information
Session Type: ACR Abstract Session
Session Time: 4:30PM-6:00PM
Background/Purpose: Identifying clinically relevant patient phenotypes amidst the variability and heterogeneity of the clinical manifestations of psoriatic arthritis (PsA) is currently challenging. Using machine-learning (ML) techniques could be the first critical step towards better understanding of disease pathotypes, and support progression towards precision medicine.1 Clusters of PsA patients based on presence/absence of disease domains were identified using ML from the secukinumab FUTURE trials program.
Methods: Hierarchical clustering was performed on the composite using “1-correlation” as the dissimilarity metric and Ward’s agglomeration method for pairwise grouping; a dendrogram was used to visualize and assess the resulting groupings. Pairwise correlations were explored in various clinical domains of PsA including dactylitis, NAPSI, PASI, enthesitis, swollen or tender joints, spondylitis, and CRP measurements from >2,700 patients across five phase 3 studies with ≈425,000 data entries at baseline and were visualized using heatmaps. Mixture (of Bernoullis) Model was applied to obtain the clusters across all domains. The model assumes that two patients share a cluster only if they have similar probability of presence of symptom across all clinical domains.
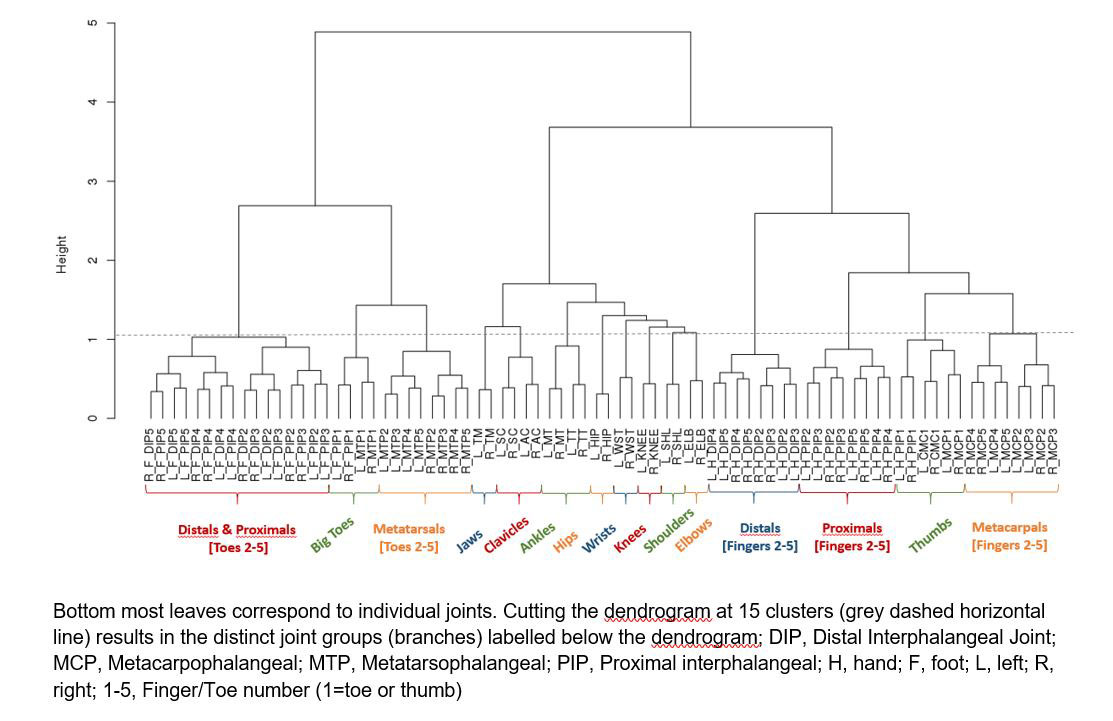
Results: The hierarchical clustering algorithm grouped the 78 individual joints into distinct and natural clusters. At higher level of the dendrogram, the algorithm grouped separately all foot, larger (jaws, clavicles, ankles, hips, wrists, knees, shoulders, elbows), and hand joints. An example for hierarchical clustering of 78 joints is shown in Figure 1. Cutting the dendrogram at 15 clusters separated all the joints into distinct groups; hand joints (distal and proximal phalanges, metacarpals and thumbs), and foot joints (distal and proximal phalanges, metatarsals and big toes). Associations of the joint clusters with other disease manifestations such as skin and nail involvement revealed distinct patient clusters, enabling to investigate differences in disease pathogenesis and treatment outcomes. For example, in one of the clusters with patients having high polyarticular burden (high number of affected joints), there was a high probability of nail involvement and psoriasis. These clusters were further refined when response to treatment over time was added in the model.
Conclusion: Machine learning methodology confirmed a natural grouping of joints in patients with psoriatic arthritis based on baseline swelling and tenderness and revealed complex correlation patterns. Additional cluster analyses including psoriatic arthritis disease manifestations and response to treatment revealed distinct patient clusters with potential clinical implications.
Reference:
- Grys BT et al., J Cell Biol. 2017; 216(1): 65-71.
To cite this abstract in AMA style:
McInnes I, Kormaksson M, Pournara E, Ligozio G, Pricop L, Nash P, Kirkham B, Reich K, Ritchlin C. Clinically Relevant Patient Clusters Identified by Machine Learning Tools in a Large Database from the Secukinumab Psoriatic Arthritis Clinical Development Program [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/clinically-relevant-patient-clusters-identified-by-machine-learning-tools-in-a-large-database-from-the-secukinumab-psoriatic-arthritis-clinical-development-program/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/clinically-relevant-patient-clusters-identified-by-machine-learning-tools-in-a-large-database-from-the-secukinumab-psoriatic-arthritis-clinical-development-program/