Session Information
Session Type: Poster Session (Sunday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Identifying pseudogout in large administrative datasets has been difficult due to lack of specific billing codes for this acute subtype of calcium pyrophosphate (CPP) crystal deposition disease. While several machine learning approaches exist to phenotype patients using electronic health record (EHR) data, they are largely validated in chronic conditions with relatively accurate billing codes. Pseudogout poses unique challenges due to its lack of specific billing codes and episodic nature. We evaluated a novel machine learning approach for classifying definite/probable pseudogout using EHR data.
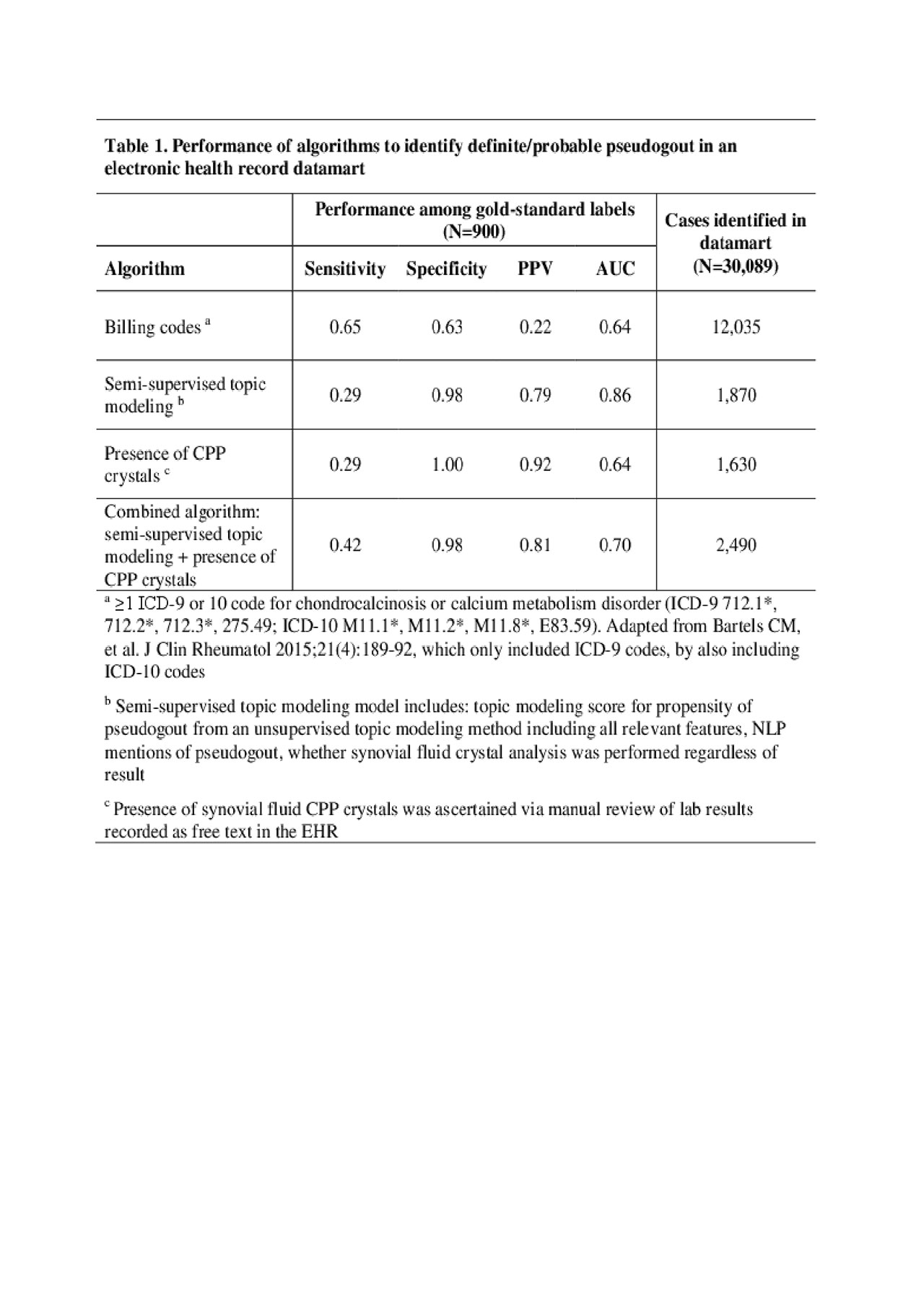
Methods: We created an EHR dataset of 30,089 patients with ≥1 relevant billing code (Table 1 footnote) or ≥2 natural language processing (NLP) mentions of pseudogout or chondrocalcinosis in narrative notes, 1990-2017. We randomly selected 900 patients for gold standard chart review to label as: (1) definite pseudogout, synovitis+synovial fluid CPP crystals; (2) probable pseudogout, synovitis+chondrocalcinosis; (3) not pseudogout. Presence of synovial fluid CPP crystals was determined by manual review of lab results recorded as free text in the EHR. To develop an algorithm for identifying definite/probable pseudogout vs. not, we applied a semi-supervised topic modeling approach; presence of CPP crystals was not included since it required manual review. The approach included the score from an unsupervised topic modeling method including all relevant features; NLP mentions of pseudogout; and whether synovial fluid crystal analysis was performed regardless of result. We created a combined algorithm including information from the semi-supervised topic modeling approach and the manually reviewed CPP crystal results. We compared algorithm accuracy and cohorts identified by: (1) billing codes, (2) presence of CPP crystals, (3) the combined algorithm.
Results: Among the 900 subjects, 123 (13.7%) had pseudogout by chart review (68 definite, 55 probable). Billing codes alone had a sensitivity 65% and PPV 22% for definite/probable pseudogout (Table 1). Presence of CPP crystals had a sensitivity 29% and PPV 92%. Without using the CPP crystal result, the semi-supervised topic modeling algorithm had a sensitivity 29% and PPV 79%. The combined algorithm yielded a sensitivity 42% and PPV 81%. The cohort identified by the combined algorithm (n=2490) was 50% larger than that identified by presence of CPP crystals (n=1630); the latter only captured patients with definite pseudogout and did not identify patients with probable pseudogout. Table 2 demonstrates important differences between cohorts identified via billing codes vs. the combined algorithm, and similarities between cohorts identified by the presence of CPP crystals vs. the combined algorithm.
Conclusion: For pseudogout, a condition without a specific billing code, combining NLP and machine learning methods with synovial fluid CPP crystal lab results yielded an algorithm that significantly boosted PPV compared to billing codes alone, with modest sensitivity. This balance allows classification of a large pseudogout cohort for future research.
To cite this abstract in AMA style:
Tedeschi S, Cai T, He Z, Ahuja Y, Hong C, Yates K, Dahal K, Xu C, Lyu H, Yoshida K, Solomon D, Cai T, Liao K. Classifying Pseudogout Using Machine Learning Approaches with Electronic Health Record Data [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/classifying-pseudogout-using-machine-learning-approaches-with-electronic-health-record-data/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/classifying-pseudogout-using-machine-learning-approaches-with-electronic-health-record-data/