Session Information
Session Type: ACR Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: The treatment of rheumatic diseases can be both expensive and ineffective with up to 1/3 of patient’s failing to respond to current treatments. There is therefore a need to identify new effective treatments and to target the best treatment to individual patients. Although genetic studies have been successful in identifying common variation associated with disease susceptibility, a large proportion of these lie outside traditional protein-coding regions. Many show enhancer activity but it is often unclear which gene(s) they regulate and how they contribute to disease. Chromatin folding brings linearly distant areas of the genome, such as promoters and enhancers, into close proximity, driving gene expression. Capture Hi-C (CHi-C) is a new method which interrogates these interactions in a high-throughput, high-resolution manner, linking implicated enhancers to causal genes. Utilising our existing CHi-C data on 3 rheumatic diseases, RA, JIA and PsA, targeting all known genetic associations, we explored the potential of this interaction data to identify potentially causal genes that are targets for existing drugs, which could be repositioned for use in these diseases.
Methods: Chromatin interaction data for T- and B-cells in the 3 diseases was re-called using CHiCAGO v2 using a score cut-off of ≥ 5. Interactions between disease regions and gene promoters were identified using BEDTOOLS v2.21.0 and intersected with known drug targets from DrugBank v4.5.0. Existing treatments for each disease were identified by the presence of the relevant name in the ‘indication’ field.
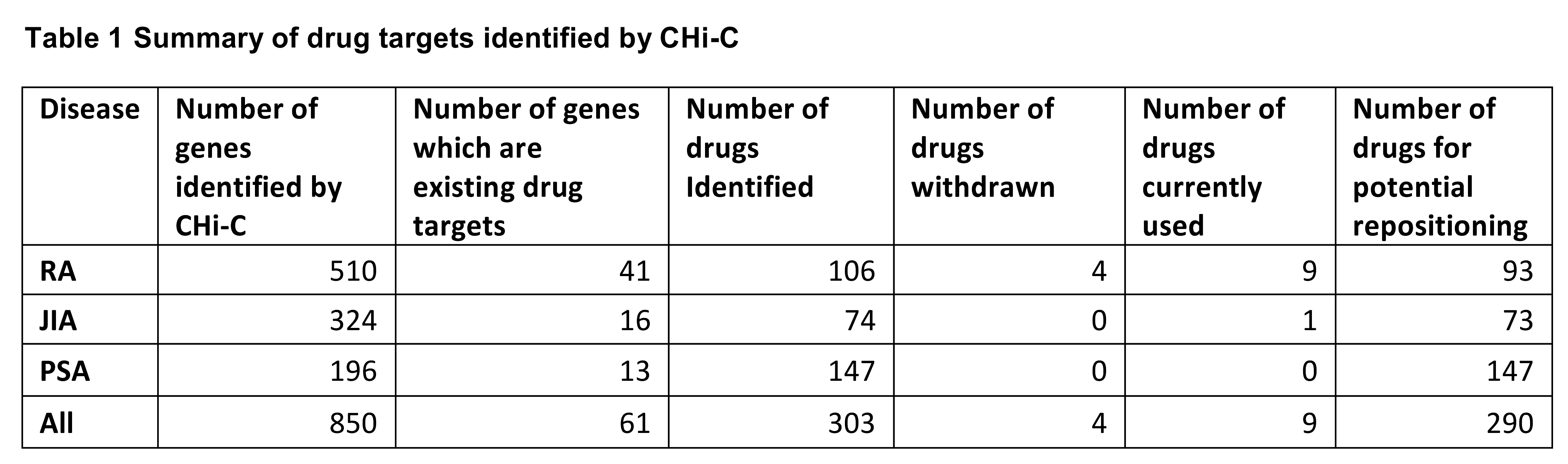
Results: Overall 850 genes were identified as interacting with a rheumatic disease associated region. Of these, 61 are existing drug targets (303 drugs) (Table 1) and 9 are existing therapies used in the treatment of disease, primarily RA.
Conclusion: Our study identifies genes which are implicated in disease, are the target of existing drugs and offer the potential for drug repositioning. Of the potential drugs identified for RA, 14 are used in the treatment of various cancers particularly leukemias and 11 are used in the treatment of diabetes and multiple sclerosis. Interestingly, 17 potential drugs identified for PsA are used in the treatment of schizophrenia and 11 in the treatment of hypertension. This data shows a novel insight into how functional annotation of genetic associations in rheumatic diseases can provide gene targets for re-positioned therapies.
To cite this abstract in AMA style:
Martin P, McGovern A, Duffus K, Yarwood A, Barton A, Worthington J, Eyre S, Orozco G. Chromatin Interactions Reveal Novel Gene Targets for Drug Repositioning in Rheumatic Diseases [abstract]. Arthritis Rheumatol. 2016; 68 (suppl 10). https://acrabstracts.org/abstract/chromatin-interactions-reveal-novel-gene-targets-for-drug-repositioning-in-rheumatic-diseases/. Accessed .« Back to 2016 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/chromatin-interactions-reveal-novel-gene-targets-for-drug-repositioning-in-rheumatic-diseases/