Session Information
Date: Sunday, October 21, 2018
Title: Rheumatoid Arthritis – Treatments Poster I: Strategy and Epidemiology
Session Type: ACR Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: Epigenetic modifications including DNA methylation are implicated in the development and progression of autoimmune diseases, such as rheumatoid arthritis [MIM 180300]. Evidence indicates that exogenous exposures, such as smoking, diet, and medications can change DNA methylation and that changes in DNA methylation contribute to immune cell autoreactivity. Methotrexate (MTX) and anti-TNF agents (a-TNFs) are effective treatments for RA. It is believed they reverse epigenetic modifications that cause T-cell autoreactivity, improving RA symptoms among some patients. Changes in DNA methylation associated with response to these treatments are potential biomarkers for prediction of treatment response.
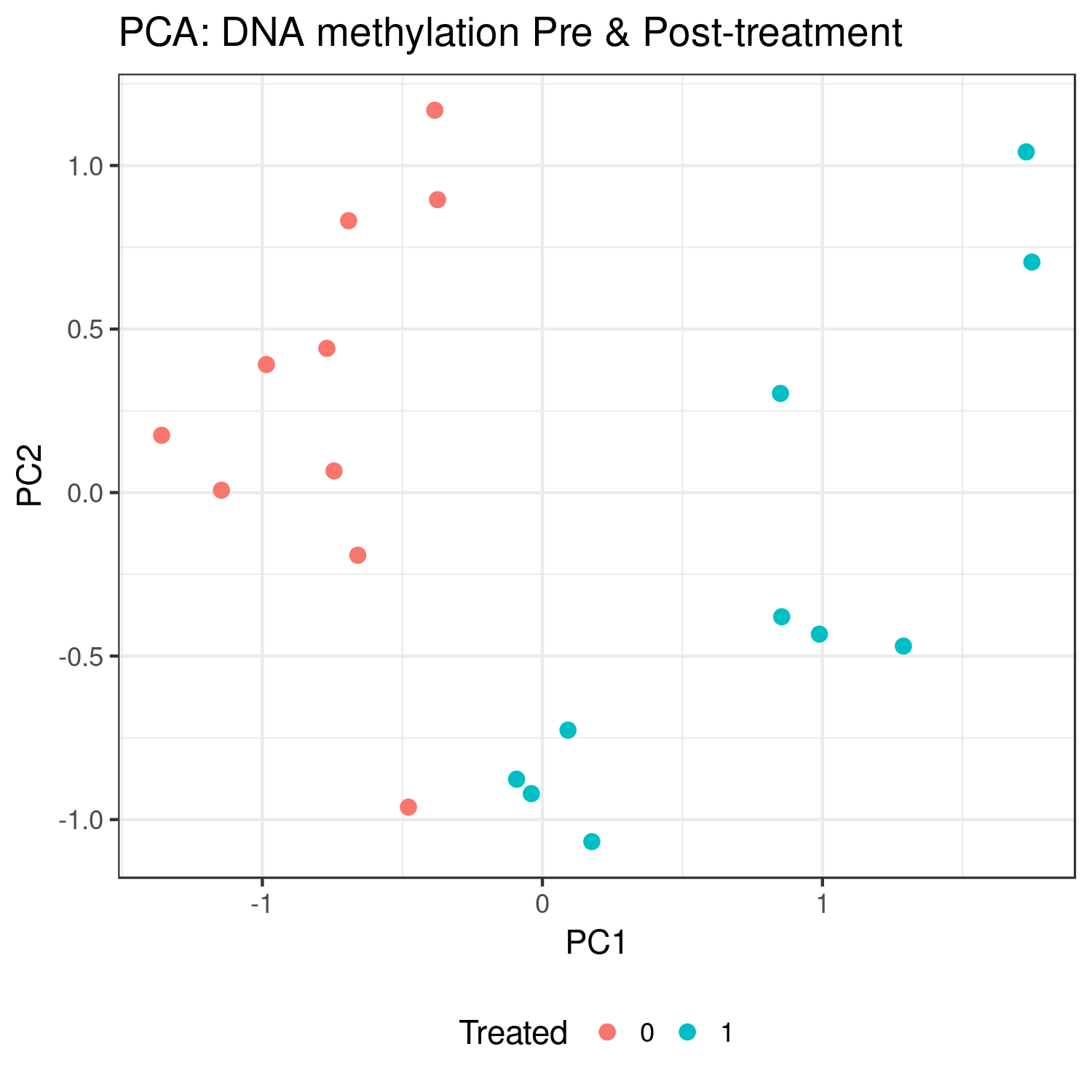
Methods: We conducted a study to identify DNA methylation profiles that may serve as biomarkers of response to treatment with MTX and a-TNFs for RA. Blood samples, clinical data, and disease severity scores (DSS) were collected from 30 treatment naive individuals with RA at baseline and after 3-6 months of treatment with: MTX (n=10), a-TNFs (n=10), or MTX and a-TNFs (n=10). DSS included the Simple Disease Activity Index, Clinical Disease Activity Index, and the C-reactive protein and Erythrocyte Sedimentation Rate Disease Activity Scores. Genome-wide methylation profiles were generated using Illumina’s Infinium Human Methylation EPIC BeadChip from PBMC samples. Quantile normalization and background subtraction with dye-bias normalization were performed using minfi and CpGs with high detection p-values and cross-reactive probes, and CpGs measuring SNPs were excluded. Treatment response was defined as difference in DSS from baseline to post treatment measurements. Differentially methylated positions (DMPs) and differentially methylated regions (DMRs) associated with, i) treatment, and ii) treatment response, were identified with limma and bumphunter, respectively, using surrogate variables to adjust for blood cell proportions, batch effects, and genetic ancestry. Next, due to small sample size, baseline methylation levels in the top 1000 DMPs and 100 DMRs (by p-value, no multiple testing correction) identified in pre- and post-treatment analyses were used to predict observed treatment response. Principal component analysis (PCA) was done with the top DMPs and DMRs to identify treatment and treatment response clusters.
Results: Preliminary results found 38 DMPs associated with treatment (p < 0.0001), 28 of which were hypomethylated. PCA found distinct clusters for study subjects pre- and post-treatment. Additionally, subjects clustered by treatment arm using baseline methylation only.
Conclusion: These results indicate that treatment with MTX and a-TNFs alter DNA methylation and may be used as biomarkers treatment response. Further research is needed with larger sample size.
To cite this abstract in AMA style:
Adams C, Marker K, Krueger M, Barcellos L, Criswell LA. Changes in DNA Methylation Identify Response to Treatment with Methotrexate and TNF Inhibitors Among RA Patients [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/changes-in-dna-methylation-identify-response-to-treatment-with-methotrexate-and-tnf-inhibitors-among-ra-patients/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/changes-in-dna-methylation-identify-response-to-treatment-with-methotrexate-and-tnf-inhibitors-among-ra-patients/