Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Giant cell arteritis (GCA) is a granulomatous vasculitis affecting cranial and large arteries in individuals over 50 years of age. Its relative rarity poses challenges for large-scale study. Institutional electronic medical record (EMR) datasets offer opportunities to examine GCA at scale but are often hindered by unstructured data formats. To address this, we developed a natural language processing (NLP) pipeline to classify temporal artery biopsy reports as positive or negative for arteritis.
Methods: Patients seen within our tri-site single-center institution with an ICD code consistent with GCA or polymyalgia rheumatica were identified. Temporal artery biopsy impression statements authored by pathologists were extracted and preprocessed using a custom regular expression script to remove Rich Text Formatting, standardize punctuation, and convert text to lowercase. These statements were then tokenized and input into a fine-tuned ClinicalBERT model1 (MedicalAI, Hugging Face). The dataset was split 80:20 into training and testing sets, with further subdivision of the training set for validation. The dataset used to train the model consisted of 349 negative and 137 positive samples. “Healed arteritis” in the report was considered a negative biopsy result. Data augmentation was implemented following initial training with 5 augmented impression reports added to the training dataset to enhance model performance. Discrimination performance was assessed using the area under the receiver operating characteristic curve (AUC).
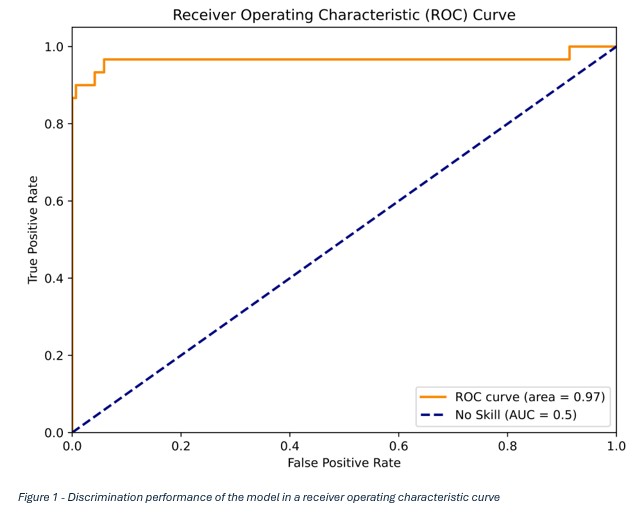
Results: The validation cohort, consisting of 320 patient records (30 positive entries and 290 negatives), was able to provide significant discrimination of positive temporal artery biopsies with an area under the curve of 0.97 (Figure 1), accuracy of 98%, precision of 96%, and recall of 87%. Of the predictions, 315/320 (98.4%) were correct. This amounted to 5 misclassifications within the dataset, which included 1 false positive and 4 false negatives (Table 1).
Conclusion: Fine-tuned language models trained on clinical text can accurately classify pathology reports, offering a scalable approach to identifying specific diagnoses within large EMR datasets. This method improves the accuracy and utility of cohort identification for rare conditions such as GCA, supporting future research efforts.References:1. 1. K. Huang, J. Altosaar, R. Ranganath, ClinicalBERT: Modeling Clinical Notes and Predicting Hospital Readmission, in: CHIL ’20 Workshop: ACM Conference on Health, Inference, and Learning 2020, Toronto, 2020.
To cite this abstract in AMA style:
Sullivan M, Rosen J, Grilli C, Warrington K, Ortega V. Capturing Patient Cohorts: A Temporal Arteritis Classifier [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/capturing-patient-cohorts-a-temporal-arteritis-classifier/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/capturing-patient-cohorts-a-temporal-arteritis-classifier/

.jpg)