Session Information
Session Type: Poster Session (Sunday)
Session Time: 9:00AM-11:00AM
Background/Purpose: The manifestations of systemic lupus erythematosus (SLE) can be divided into categories according to a recently proposed model: a category of classic active autoimmune-driven manifestations that cause organ damage (Type 1) and a second category of fatigue, myalgia, mood disturbance, and cognitive dysfunction (Type 2); whereas Type 1 signs are treated with immunosuppression, Type 2 symptoms remain resistant to immunosuppression. Distinguishing between Type 1 and 2 SLE can be challenging, especially for the non-rheumatologist. We sought to determine whether transcriptomic differences distinguish between these categories of lupus.
Methods: Using a book-ended approach to avoid clinical overlap in the populations, we identified two cohorts of women with lupus. The first consisted of 10 women with Type 1 disease, defined by a SLEDAI ≥6 and a fibromyalgia severity score (FSS) < 9. A second cohort of 10 women had Type 2 lupus with a SLEDAI of 0 and an FSS > 9. Whole blood was collected in PAXgene Blood RNA tubes and whole transcriptome RNA sequencing was performed. Hierarchical clustering and limma differential expression (DE) analysis were used to compare women of African heritage in the Type 1 and Type 2 cohorts to limit heterogeneity by race. Gene set variation analysis (GSVA) was carried out on all Type 1 and Type 2 lupus patients using modules of co-expressed genes obtained from DE analysis. Clinical characteristics were compared by Fisher’s Exact test.
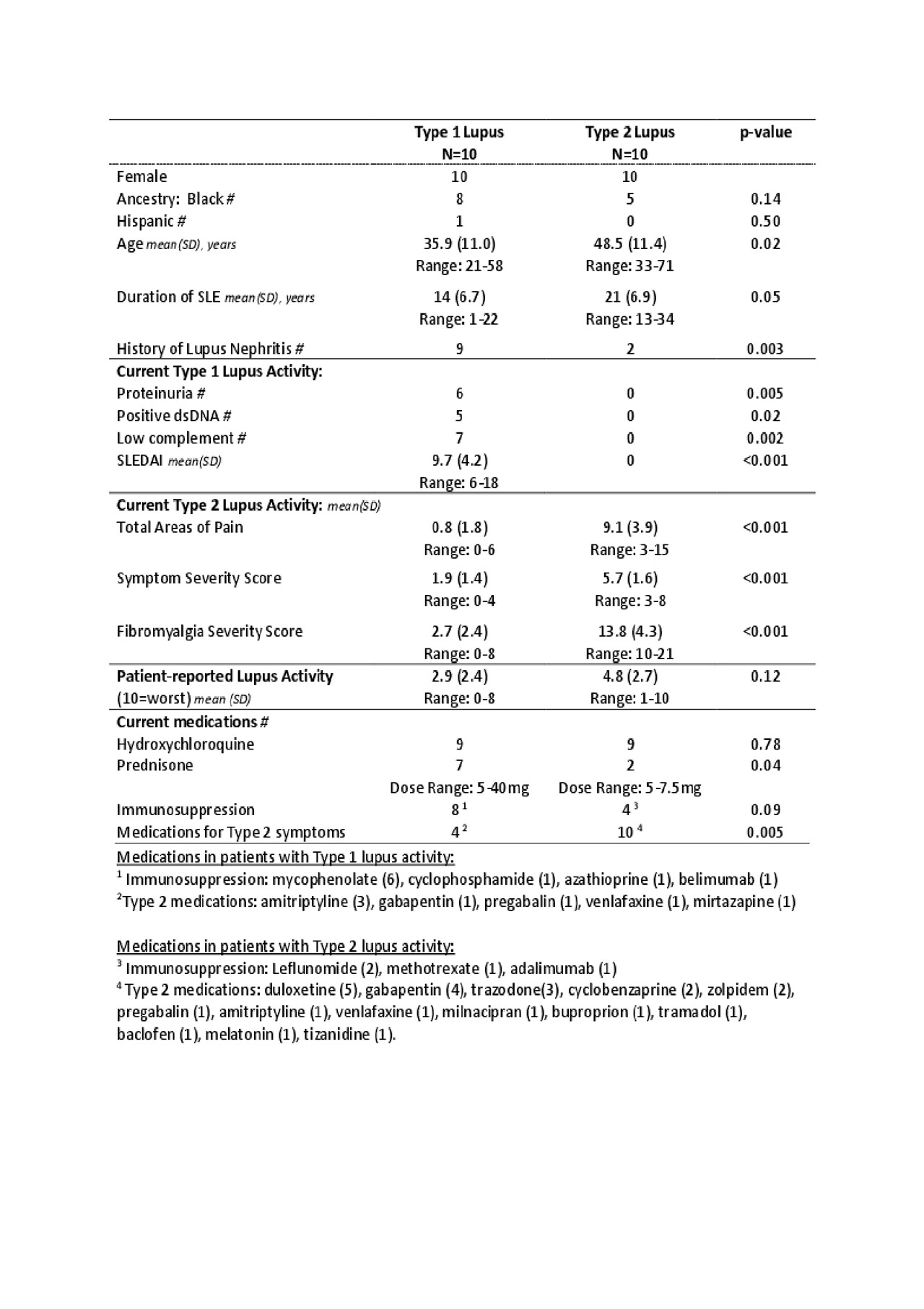
Results: Women with symptomatic Type 1 lupus were younger and had a shorter disease duration; most had prior and current nephritis; 5 had elevated dsDNA antibodies, and 7 had low complement (Table 1). The average SLEDAI was 9.8 (range 6-18). Of women with Type 2 lupus, 20% had a history of lupus nephritis and none had current proteinuria. Women with Type 2 lupus had more areas of pain, symptoms of fatigue, brain fog, and non-restorative sleep. Interestingly, the patient assessment of lupus activity did not differ between the groups. The large majority of women in both groups were taking hydroxychloroquine. More women with Type 1 lupus took immunosuppressants and corticosteroids whereas more women with Type 2 lupus took medications for pain and depression.
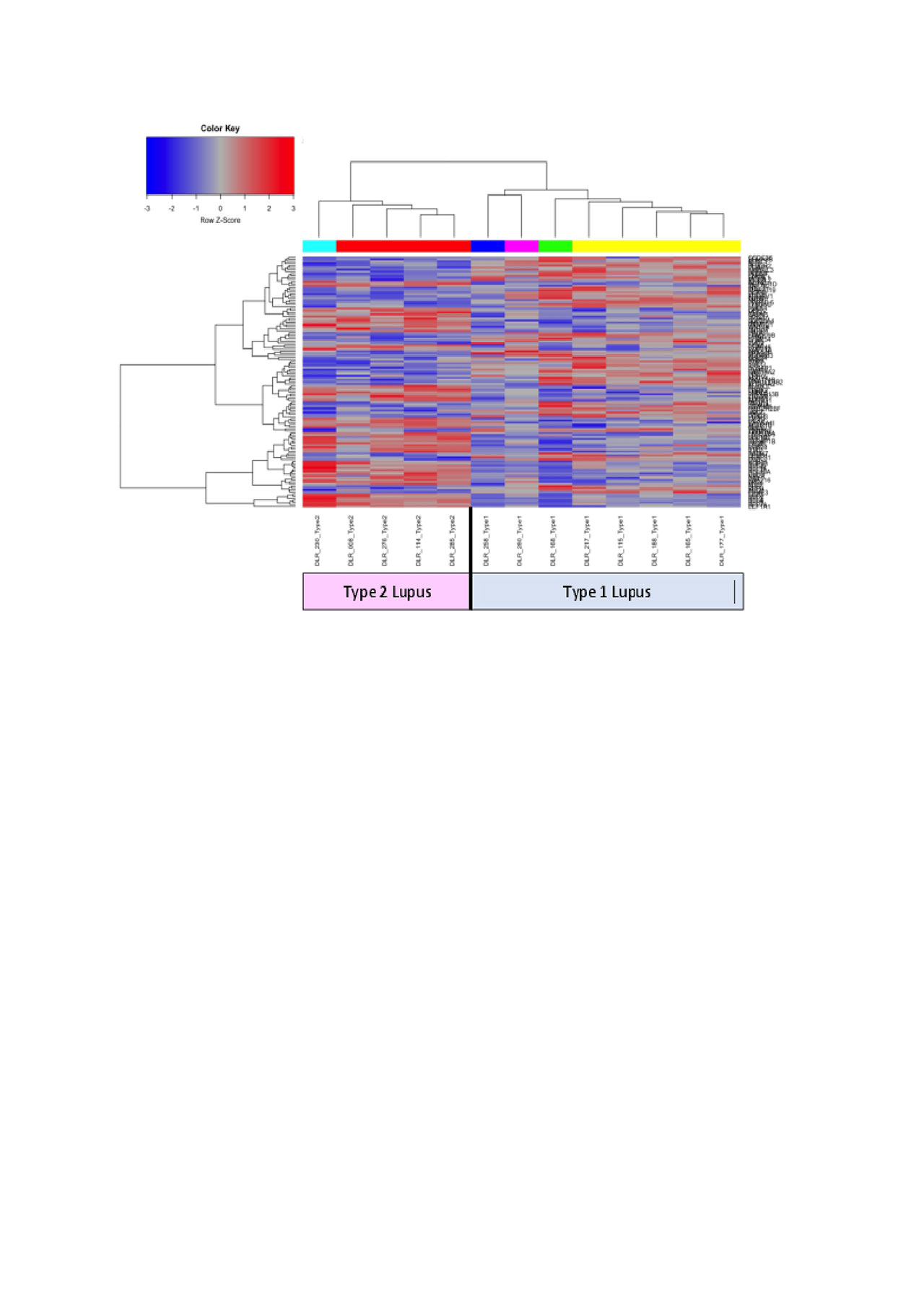
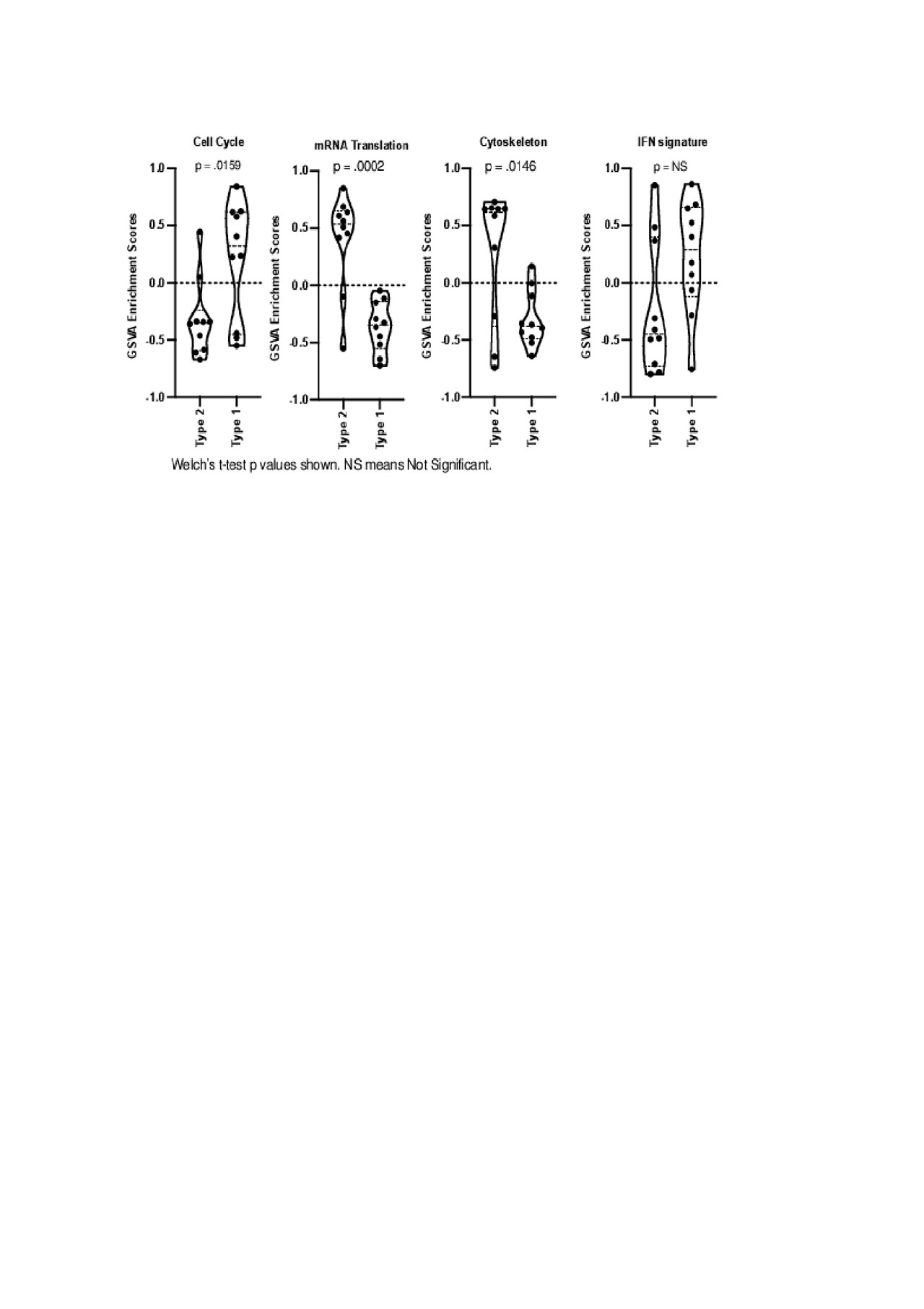
DE analysis determined distinct hierarchical clustering between Type 1 and Type 2 lupus (Figure 1). Biologically-informed gene clustering identified transcripts for interferon signature genes, low density granulocytes, cell cycle genes, and SDC1, a marker of plasma cells, as increased in Type I lupus; transcripts associated with mRNA translation and the cytoskeleton were increased in Type 2 lupus. GSVA using these signatures demonstrated that the cell cycle signature was significantly (p < .05) enriched in Type 1 lupus and mRNA translation and the cytoskeleton were significantly (p < .05) enriched in Type 2 lupus (Figure 2).
Conclusion: Our new conceptual model characterizes lupus in terms of two categories: Type 1, with immunologic activity and organ involvement, and Type 2, with minimal immunologic activity but debilitating chronic symptoms. Transcriptomic analysis provides evidence that these categories are biologically distinct and can be distinguished by uniquely aberrant molecular pathways.
To cite this abstract in AMA style:
Clowse M, Rogers J, Eudy A, Criscione-Schreiber L, Doss J, Sadun R, Sun K, McClain M, Tsalik E, Woods C, Pisetsky D, Bachali P, Grammer A, Catalina M, Lipsky P. Biologic Differences Between Type 1 and 2 Lupus [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/biologic-differences-between-type-1-and-2-lupus/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/biologic-differences-between-type-1-and-2-lupus/