Session Information
Session Type: Poster Session (Tuesday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Skin inflammation can drive systemic disease in SLE, thus it is essential to understand the key regulators of the aberrant inflammatory response in lupus skin. We have previously identified BATF2 as a transcription factor that is highly overexpressed in lupus keratinocytes following type I IFN exposure. The objective of this study was to determine the role of BATF2 in regulating keratinocyte responses to type I and type II interferons.
Methods: After generating BATF2 knockouts (BATF2 -/-) in a human keratinocyte cell line (N/TERTs) using CRISPR/Cas9 technology, we treated both BATF2 -/- and control N/TERTs (CTL) with either media or 1000 units/milliliter of IFN alpha (α) or gamma (γ) for 6 hours. We subsequently isolated RNA from biological triplicates of each treatment condition and completed Illumina HiSeq analysis through the University of Michigan Sequencing Core. We performed RNA sequencing (RNA-seq) analysis to identify differentially expressed genes (DEGs; p-adjusted ≤ 10% and log2|FC| ≥ 1) and differences in transcriptional profiles between treatment conditions. We then utilized Ingenuity Pathway Analysis (IPA) to highlight enriched biological pathways within regulated genes and Genomatix Pathway System (GePS) to display transcriptional targets of regulated genes. To validate our RNA-seq findings, we performed RT-PCR of candidate genes and BATF2 immunostaining of cutaneous lupus (CLE) biopsies.
Results: RNA-seq analysis revealed an overall higher number of DEGs in CTL compared to BATF2 -/- with both type I and II interferon stimulation (905 and 1542 genes versus 536 and 947, respectively). DEGs unique to CTL upon IFNα stimulation included IFNK, IL-16 and HLA-DRB1, and with IFNγ, MX2, TNF, TLR5 and CASP5. Of identified DEGs, 408 were common to CTL and BATF2 -/- with IFNα and 744 with IFNγ stimulation, including the majority of IFN-responsive genes. Within common DEGs upon IFNα and γ stimulation, 127 and 215 genes were 1.5-fold increased or decreased in BATF2 -/- relative to CTL. IFN-responsive genes less expressed in BATF2 -/- with IFN stimulation compared to CTL included CXCL10, OASL, CCL5, IFIT1/2/3, CASP1, IFI30 and ICAM1, with some genes common and unique to IFNα and γ stimulation (Figure 1). Not all IFN-responsive genes were differentially regulated by BATF2, and some were even more expressed in BATF2 -/- (CXCL9, MX1). Transcription factor analysis showed enrichment for key genes in the IFN response with a potential binding site for BATF2 in their promoter, including CXCL9/10, IRF1, JAK2, MX1 and TLR2, many of which were identified as differentially regulated by BATF2. To validate RNA-seq data, we confirmed candidate differentially regulated genes between BATF2 -/- and CTL using RT-PCR, notably decreased IFNK expression in BATF2 -/-, and demonstrated increased BATF2 expression in CLE biopsies compared to healthy control skin.
Conclusion: BATF2 -/- keratinocytes respond similarly to CTL upon IFN stimulation with respect to expression of many IFN-responsive genes; however, critical genes dysregulated in CLE lesions, including CXCL10, CCL5 and CASP1 require BATF2 for peak induction by IFNs. Thus, BATF2 may be a specific target for blocking IFN-induced inflammatory responses in SLE skin.
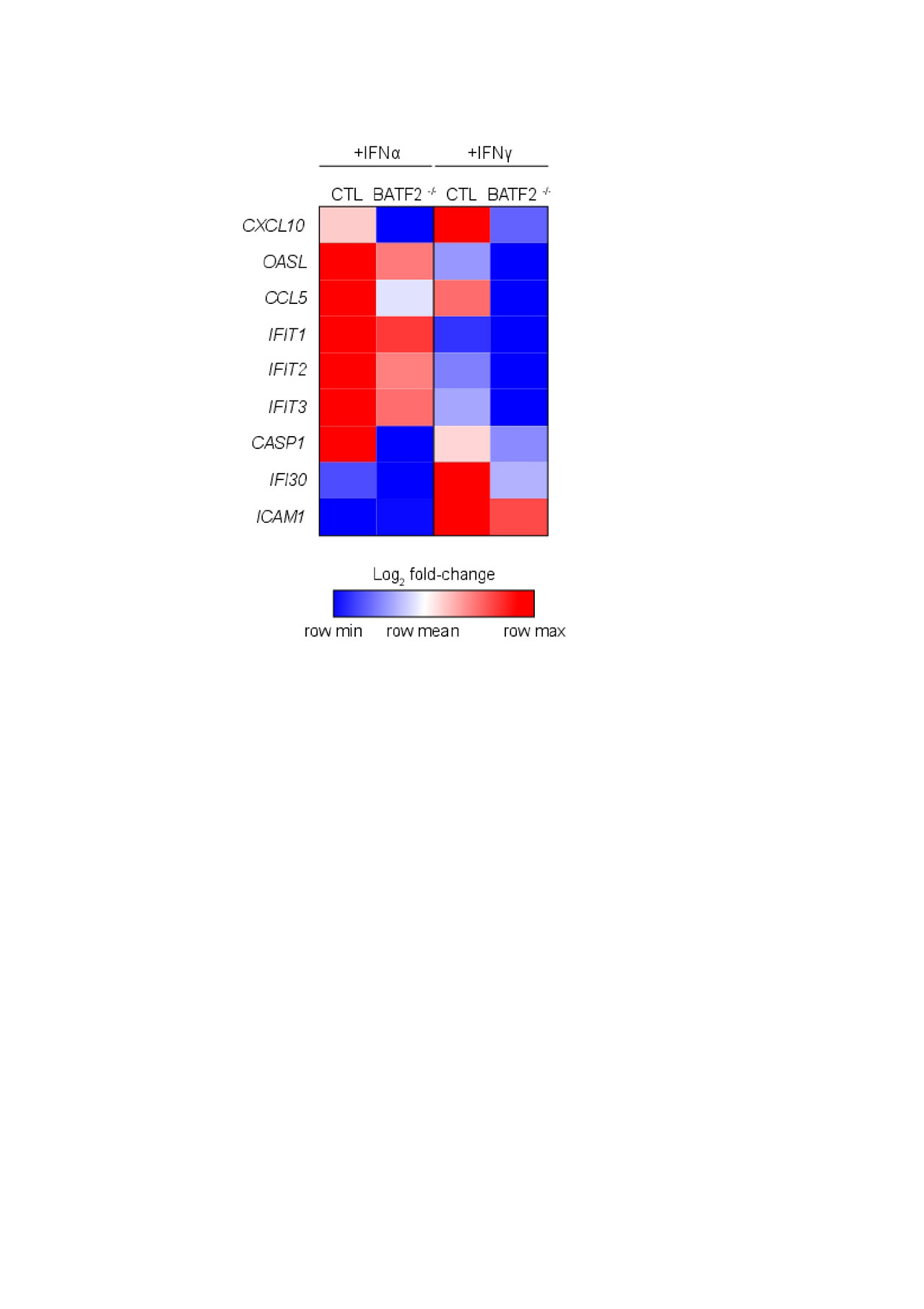
Heatmap_for BATF2_ACR abstract_FINAL
To cite this abstract in AMA style:
Turnier J, Berthier C, Tsoi L, Barnes D, Hile G, Reed T, Liu J, Gudjonsson J, Kahlenberg J. BATF2 Contributes to Interferon Dysregulation in SLE Keratinocytes [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/batf2-contributes-to-interferon-dysregulation-in-sle-keratinocytes/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/batf2-contributes-to-interferon-dysregulation-in-sle-keratinocytes/
