Session Information
Date: Tuesday, November 12, 2019
Title: Epidemiology & Public Health Poster III: OA, Gout, & Other Diseases
Session Type: Poster Session (Tuesday)
Session Time: 9:00AM-11:00AM
Background/Purpose: The use of Electronic Medical Records (EMR) for research purposes has led to an increasing interest in Natural Language Processing (NLP) for text classification. Little preparation is required as the NLP-methods are capable of automatically interpreting the data. Classification is often accomplished with naïve word-matching. An alternative to word-matching is the usage of a Machine Learning (ML) classifier. Rather than providing a set of patterns, an ML-model only requires the outcome and then formulates the patterns itself. The purpose of this study is to build a reliable classifier with machine learning techniques that can identify the Rheumatoid Arthritis (RA) cases based on the provided EMR entry.
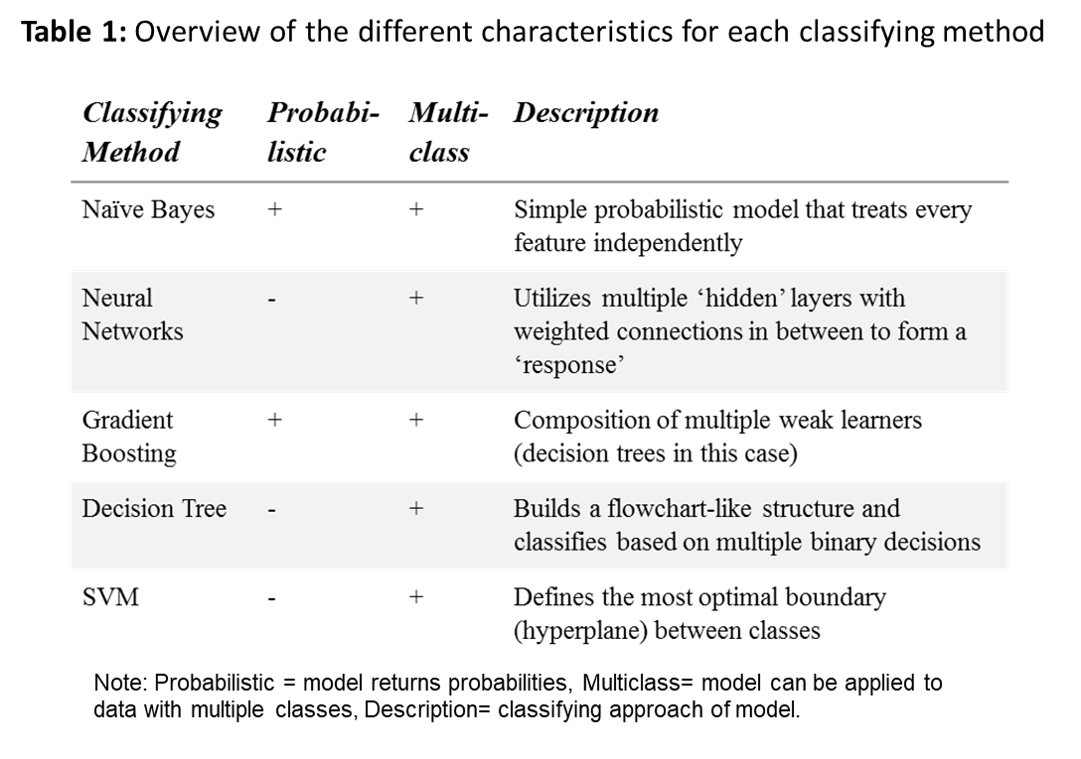
Methods: Data was acquired from the HiX-EMR database consisting of 2,771 patients that visited the rheumatology outpatient clinic from the Leiden University Medical Centre between 2007 and 2018. This database featured a total of 38,216 entries. The first entry (if available) was selected per patient for annotation, resulting in a total of 1,361 entries. The annotated sample was then randomly split into an equally sized training and test set. Both sets were preprocessed and then classified with the following methods: naïve word-matching, Naive Bayes (NB), Decision Tree, Gradient Boosting (GB), Neural Networks and Support Vector Machines (SVM), see table 1 for more information. Default Scikit-learn implementations2 were used to create the models, except for the word-matching model of which the classification is based on the presence of RA-defining strings like ‘Reumatoide Artritis’.
Finally, the performance of the models was evaluated with a receiver operating characteristic (ROC) curve analysis via the pROC package3. The Delong test was used to assess the 95% confidence intervals (CI) and to determine the difference between the performance of the word-matching method and the ML-models.
Results: The naïve word-matching approach resulted in a high area under the curve (AUC=0.76). Likewise, the ML-models resulted in relatively high AUC-scores as well: NB=0.83, SVM=0.91, Neural Networks=0.92 and the GB-method with a 0.94. The Decision Tree showed the worst performance with an AUC-ROC of only 0.51. In comparison to the naïve word-matching ROC-curve, all the ML-models showed a significant difference: Decision Tree (p< 2.2e-16; CI=0.49-0.56), NB (p= 4.4e-3; CI=0.80-0.86), Neural Networks (p< 2.2e-16; CI=0.90-0.94), GB (p< 2.2e-16; CI=0.92-0.96) and the SVM (p=4.0e-16; CI=0.89-0.93).
Conclusion: The Gradient Boosting, Neural Networks, SVM and Naïve Bayes models all showcased a significantly better performance than the Naïve word-matching algorithm, which establishes these ML-methods as an efficient approach for data extraction from EMR.
To cite this abstract in AMA style:
Maarseveen T, Huizinga T, Reinders M, van den Akker E, Knevel R. Automated Diagnosis Extraction from Electronic Medical Records with Machine Learning Classifiers [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/automated-diagnosis-extraction-from-electronic-medical-records-with-machine-learning-classifiers/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/automated-diagnosis-extraction-from-electronic-medical-records-with-machine-learning-classifiers/