Session Information
Session Type: Poster Session B
Session Time: 9:00AM-10:30AM
Background/Purpose: Patients with rheumatic skin diseases such as cutaneous lupus erythematosus (CLE) and systemic sclerosis (SSc) can be classified from individuals with healthy skin using the enrichment of specific molecular signatures [1]. However, as patient heterogeneity is a well-known feature of these diseases, it is important to ascertain whether there are distinct patient molecular phenotypes (endotypes). Moreover, identifying specific disease endotypes from analysis of peripheral blood might increase the capacity to recognize patient endotypes.
Methods: Gene expression data derived from publicly available lesional skin biopsies of CLE, specifically discoid lupus erythematosus (DLE), the most severe CLE subtype, and SSc patients was analyzed using gene set variation analysis (GSVA) of informative gene modules. Paired blood samples were also analyzed when available. K-means clustering was then applied to the GSVA enrichment scores to identify molecular endotypes in both skin diseases.
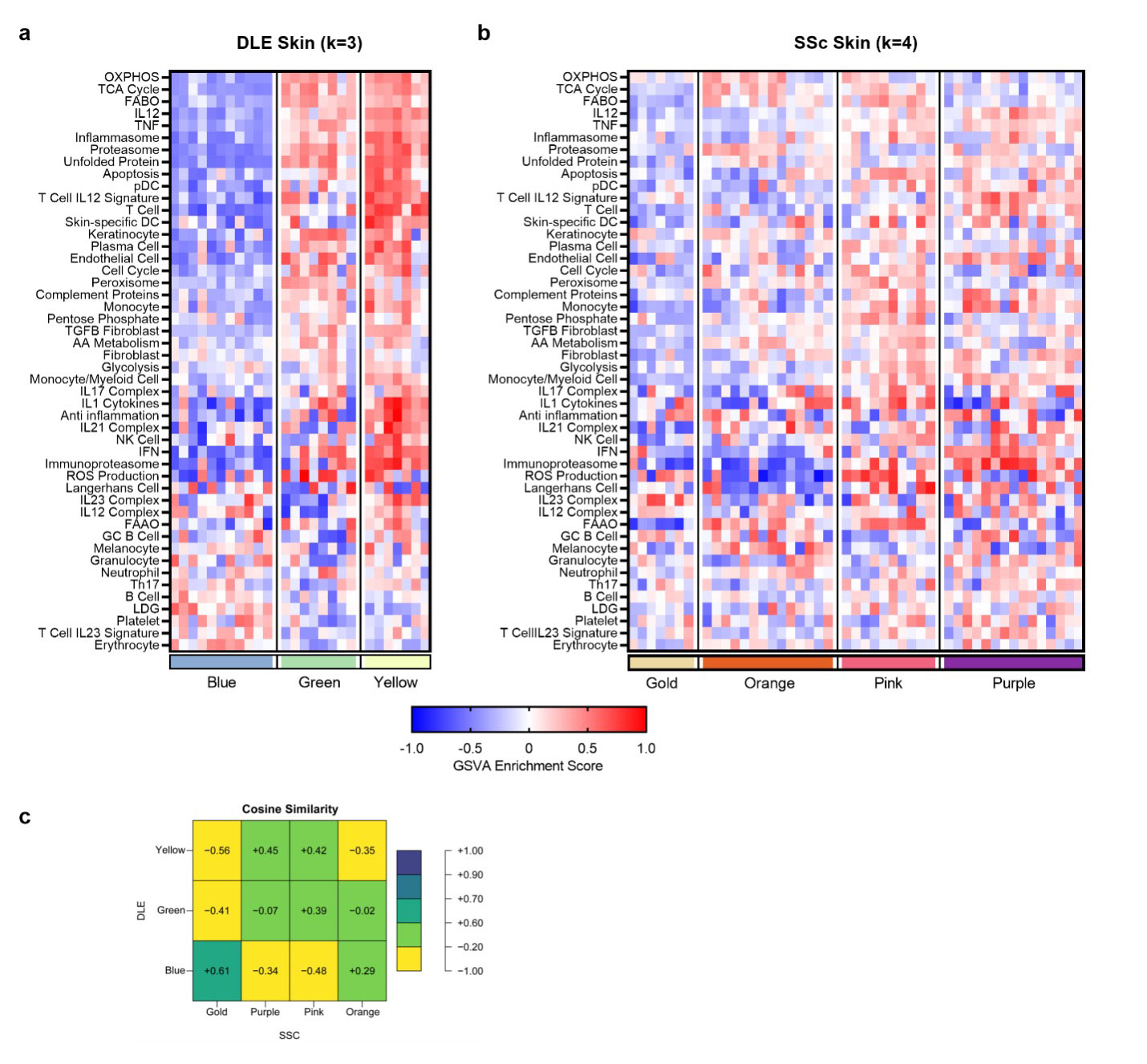
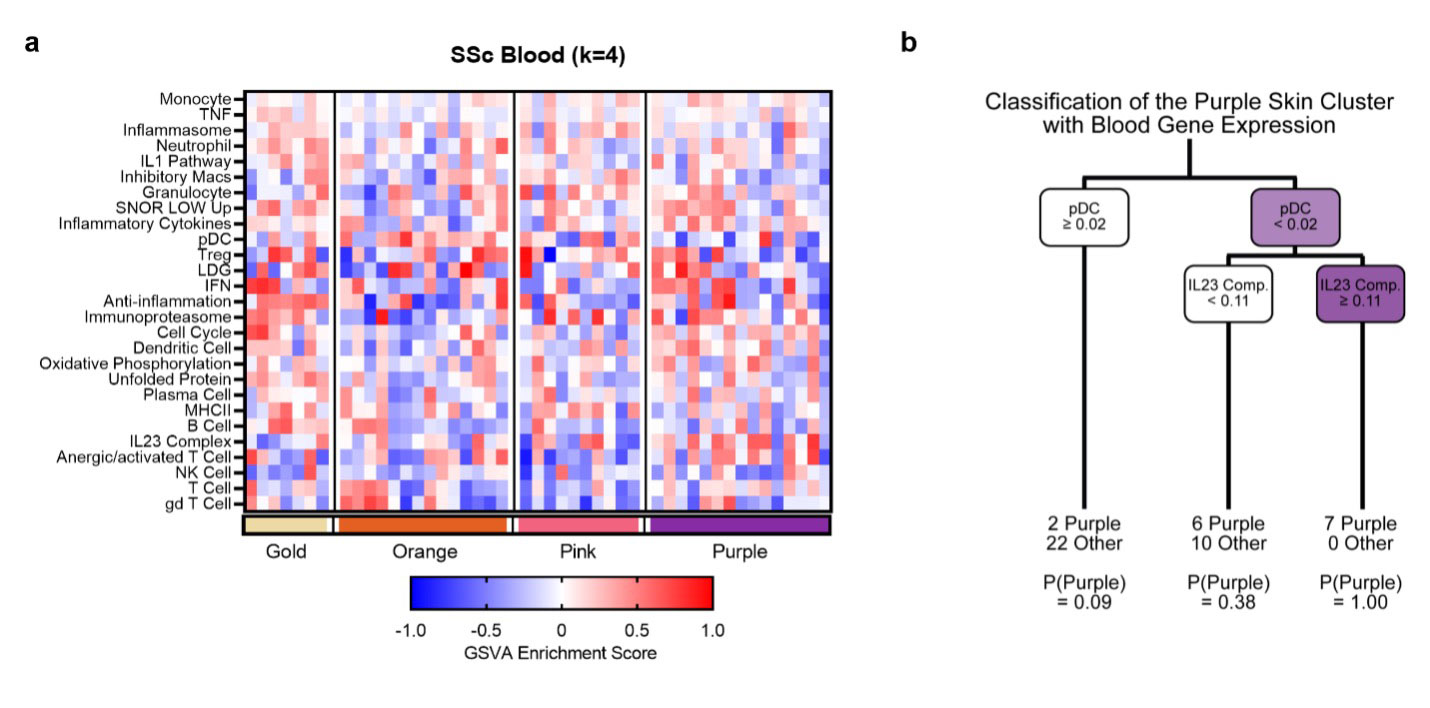
Results: In DLE, k-means clustering revealed three subsets with distinct features (Fig. 1a). The least abnormal DLE cluster (blue) was characterized by minimal changes, including higher B cell GSVA scores, whereas the other two clusters (green and yellow) had markedly increased inflammatory cell and pathway gene expression, with most modules being enriched by a greater magnitude in yellow. In SSc, four endotypes emerged from k-means clustering (Fig. 1b). The first cluster (gold) was the least abnormal, with unchanged cell and metabolic signatures. The second cluster (orange) was characterized by increased melanocyte transcripts and altered metabolic signature expression. The third cluster (pink) exhibited increased myeloid, NK, T, and plasma cell signatures as well as increased glycolysis, pentose phosphate, and apoptosis signatures. The most severe SSc endotype (purple) was comprised of increased myeloid cell, T cell, and fibroblast signatures, and altered metabolism expression. Both the pink and purple clusters were enriched in cytokine signaling modules such as IL12 and TNF. Comparison of the three DLE endotypes with the four SSc endotypes revealed some overlap between the least severe clusters (blue and gold) of each disease (Fig. 1c). Finally, we examined the ability to predict SSc skin endotypes from the blood of paired samples. Blood was analyzed for enrichment of blood-specific gene signatures and then the blood samples were grouped into the SSc skin-identified clusters (Fig. 2a). The most severe SSc skin endotype could be predicted by Classification and Regression Tree (CART) analysis of blood, a negative plasmacytoid dendritic cell (pDC) GSVA score in the blood predicted the purple cluster (Fig. 2b).
Conclusion: Both DLE and SSc skin exhibit distinct subsets based upon their molecular profile (endotype). In addition, the most severe SSc skin subset can be identified from blood gene expression. Identifying specific molecular endotypes of patients with inflammatory skin disease may facilitate matching individual patients with effective therapies.
To cite this abstract in AMA style:
Shrotri S, Kingsmore Allison K, Bachali P, Grammer A, Lipsky P. Analysis of the Transcriptomic Profiles of Rheumatic Skin Diseases Reveals Disease-specific Endotypes [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/analysis-of-the-transcriptomic-profiles-of-rheumatic-skin-diseases-reveals-disease-specific-endotypes/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/analysis-of-the-transcriptomic-profiles-of-rheumatic-skin-diseases-reveals-disease-specific-endotypes/