Session Information
Session Type: Abstract Submissions (ACR)
Background/Purpose: Genome wide association studies (GWAS) have identified loci associated with complex traits, and the current challenge is to glean biological insights from these findings. A GWAS in 110,347 samples discovered ~30 loci associated with serum urate; however, only two loci were associated with gout in 2,115 cases (SLC2A9, ABCG2)1. The strongest effects lie in renal urate transporter loci, with additional loci thought to include metabolic genes. Here we systematically assess polygenic effects of urate transport, metabolic and immune-related genes, excluding known associated loci, in both serum urate and gout.
Methods: GWAS results (2.2M HapMap2-imputed SNPs) for serum urate (110,347 individuals) and for gout case/control status (2,115 cases in 14 studies), were utilized to construct polygenic scores. After removal of the 31 significant urate GWAS loci (1Mb each), non-overlapping genomic compartments were defined by SNPs in proximity (+- 50Kb) to genes involved in 1. Urate transport: ABCG and SLC family members of urate-associated transporters and their direct connections in Inweb and String network databases2 (348 genes), 2. Urate metabolism: purine metabolism and glycolysis KEGG pathways (256 genes), 3. Immune-related: genes targeted by the Immunochip array (184 genes), 4. All other genic regions, and 5. Intergenic regions. Polygenic scores based on GWAS P-value thresholds3 were calculated in Nurses Health Study (NHS) and the Health Professionals Follow-up Study (HPFS) samples, with incident gout cases having met ACR criteria for diagnosis, and were tested in multivariate analysis with sex, age, BMI and a 31 SNP urate genetic risk score as covariates.
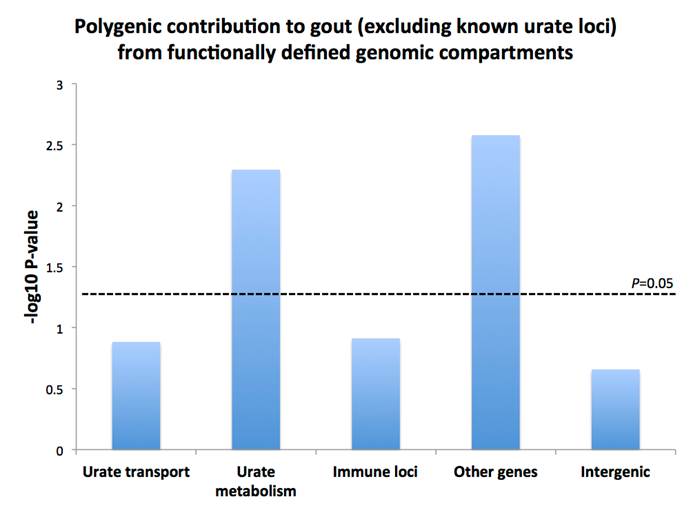
Results: Polygenic scores were significantly associated with gout in metabolic genes (n=797 SNPs with PGWAS<10-2; P=0.005) and other genic regions (n=570,599 SNPs with PGWAS<0.5; P=0.001), with similar results for association with serum urate (P=0.0025 and P=0.007 for metabolic and other genes respectively). The immune gene compartment was not significant in gout (P>0.12). The urate transporter compartment (excluding known significantly associated urate transporter loci) was not significant in gout or urate (P>0.13 and P=0.20, respectively).
Conclusion: Our results demonstrate an important role for metabolism in both serum urate and risk of gout, with genes outside of the defined compartments also contributing. Excluding known loci, we find no additional evidence for polygenic variation in urate transport or immune-related functions contributing to serum urate or gout. Genomic compartments defined on function can provide additional discoveries and further biological insights into the etiologies of urate and gout.
1. Köttgen, Nat Genet 45: 145 (2013)
2. Lage, Nat Biotechnol 25: 309 (2007); Franceschini, Nucleic Acids Res 41: D808 (2013)
3. Purcell, Nature 460: 748 (2009)
Disclosure:
E. A. Stahl,
None;
T. R. Merriman,
None;
A. Dobbyn,
None;
D. B. Mount,
None;
P. Kraft,
None;
H. K. Choi,
Takeda,
5,
AstraZeneca,
5.
« Back to 2014 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/polygenic-analysis-of-transport-metabolism-and-immune-related-genomic-compartments-in-serum-urate-and-gout/