Session Information
Date: Tuesday, October 28, 2025
Title: Abstracts: Systemic Lupus Erythematosus – Etiology and Pathogenesis (2597–2602)
Session Type: Abstract Session
Session Time: 2:15PM-2:30PM
Background/Purpose: Systemic lupus erythematosus (SLE) is a complex autoimmune disease with unknown etiology. While we previously identified key gene signatures of SLE using bulk RNA-seq from 27 immune cell types (Cell 2022), we failed to identify more granular disease-relevant cell states within each cell type. Although single-cell RNA-seq (scRNA-seq) can address this problem, recent studies ended up defining ~20 coarse cell types and relied on cell-type-level pseudobulk analysis, which has hindered the discovery of new drug targets. Our in-depth investigation strategy for single-cell data, cellular and molecular fine mapping, aimed to identify new disease-relevant cell states in SLE and to perform a detailed functional characterization of them.
Methods: We applied our strategy to the largest-scale single-cell data of SLE with ~1.8 million peripheral blood mononuclear cells from 97 healthy controls and 191 SLE cases (Figure 1). We identified granular cell states that expanded in SLE (cellular fine mapping) and investigated their molecular profile using in silico and experimental approaches (molecular fine mapping).
Results: After we identified 118 granular cell states in 25 cell types with fine-grained clustering, we discovered that previously uncharacterized cell states such as FAM13A+ARID5B+ naive CD4+ T cells, ARHGAP15+CAMK4+ T cells, GZMK+GZMH+HLA-DR+ effector memory CD8+ T cells, and C1Q+ monocytes, expanded especially in clinically active SLE (covarying neighborhood analysis, Figure 2).Next, our new statistical model, which can decompose conventional pseudobulk-level differential expression into cell state abundance signature genes (quantitative change) and dysregulated signature genes (qualitative change) at single-cell level, showed that classical immunological pathways were more enriched in cell state abundance signature genes rather than dysregulated signature genes. Integrative analysis using genome-wide association studies (SLE-GWAS) with sc-linker pipeline also revealed that SLE polygenic signals were more linked to cell state abundance signature genes, suggesting that these expanded cell states can exert causal effects on SLE pathogenesis.Finally, we investigated detailed molecular profiles of these new disease-relevant cell states. For example, GZMK+GZMH+HLA-DR+ effector memory CD8+ T cells exhibited the highest expression of exhaustion signatures (PDCD1 and TIGIT) and key cytokines (IFNG and IL16, Figure 3). Integrative analysis with 137 cell surface markers identified four key surface markers of this subpopulation, which enabled us to perform in-depth functional characterization through experimental approaches such as in vitro stimulation. Furthermore, CRISPR gene knockout screening also identified several transcriptional regulator candidates of these cell states.
Conclusion: Our intensive cellular and molecular fine-mapping approach successfully pinpointed new disease-relevant cell states in SLE and their key molecules, which could be the next treatment targets.
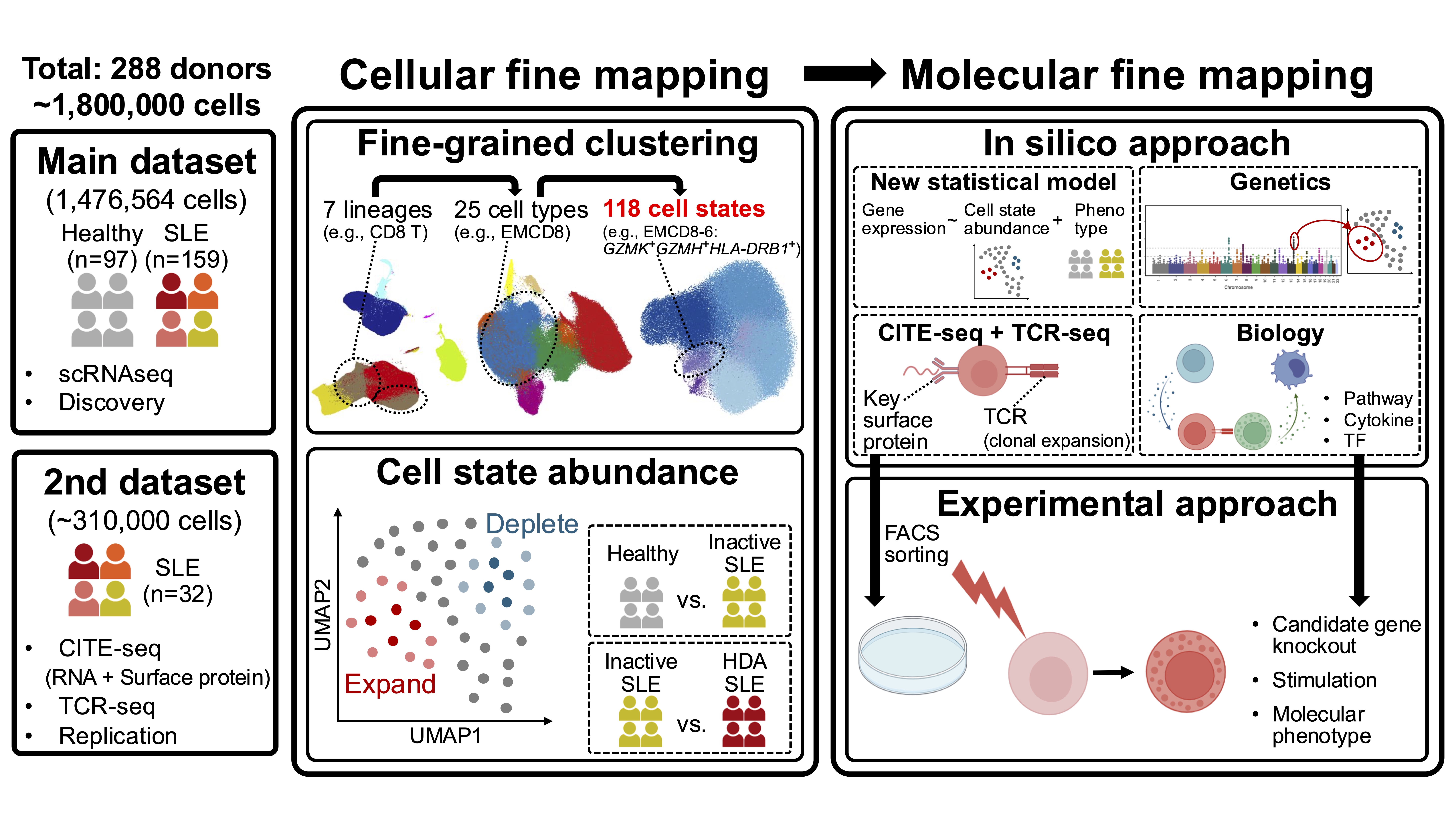 Figure 1. Overview of this study
Figure 1. Overview of this study
.jpg) Figure 2. Granular cell states that expanded in SLE
Figure 2. Granular cell states that expanded in SLE
.jpg) Figure 3. Molecular profiles in GZMK+GZMH+HLA-DR+ effector memory CD8+ T cells
Figure 3. Molecular profiles in GZMK+GZMH+HLA-DR+ effector memory CD8+ T cells
To cite this abstract in AMA style:
Nakano M, Kono M, Asahara K, Katsuyama T, Katsuyama E, Arakawa T, Kawashima T, Inokuchi H, Nishino T, Takahashi H, Natsumoto B, Hatano H, Matsumoto Y, Ishigaki K. Cellular and molecular fine mapping in single-cell data pinpoints new immunopathology of systemic lupus erythematosus [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/cellular-and-molecular-fine-mapping-in-single-cell-data-pinpoints-new-immunopathology-of-systemic-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/cellular-and-molecular-fine-mapping-in-single-cell-data-pinpoints-new-immunopathology-of-systemic-lupus-erythematosus/
