Session Information
Date: Tuesday, October 28, 2025
Title: (1830–1854) Systemic Lupus Erythematosus – Etiology and Pathogenesis Poster
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Cutaneous lupus erythematosus (CLE) is a chronic inflammatory skin disease associated with systemic lupus erythematosus (SLE) and characterized by a prominent type I interferon response. Although the mechanisms that trigger cutaneous lupus lesions remain unclear, recent therapeutic developments have highlighted a pivotal role for plasmacytoid dendritic cells (pDCs) in its disease pathogenesis.
Methods: As part of the NIH AMP-AIM-supported Lupus Omics Cutaneous Kidney Investigation Team (LOCKIT), we performed advanced multi-omics profiling using single-cell and spatial transcriptomics on paired lesional and non-lesional skin samples from 12 patients with CLE, including 11 with discoid lupus erythematosus (DLE) and 1 with subacute CLE (SCLE). CLE patients were treatment-naïve or on treatment with hydroxychloroquine only.
Results: Biopsies were obtained from scalp (n=10) and back (n=2) from 12 patients (5 Black, 7 White, 80% female). For scRNA-seq, we generated 537 million reads from over 130,000 cells. Our analysis identified two distinct pDC populations, pDC1 and pDC2 (pDC-like cells), based on expression of LILRA4 (ILT7), CLEC4C (BDC2A), LTK, GZMB, SIGLEC6, and IRF7. Both pDCs predominantly enriched in lesional skin ( >74%), with pDC1 comprised 0.06% to 4.66% of total cells and pDC2 ranged from 0% to 0.31%, while both were markedly reduced (~0.16%) in non-lesional skin. pDC1 (891 cells) expressed classical pDC markers such as GZMB, C12orf75, IRF7, SOX4, and LDLRAD4. In contrast, pDC2 (68 cells) showed lower GZMB and higher expression of PPP1R14A, LTK, and KCNK17, and clustered with the rest of the myeloid cells, suggesting a transitional myeloid state (Figure 1). Among the differentially expressed genes, CLEC4C (BDCA2) was significantly higher in pDC1 (adjusted p = 7.09 × 10⁻¹⁹, log₂FC = 2.48) compared to pDC2. While both LILRA4 (ILT7) and CLEC4C (BDCA2) were found prominently on pDCs and pDC-like cells, lower intensity of CLEC4C was seen on a larger percentage of cDC1 cells, while ILT7 was present on a small number of cDC1 cells. Macrophages and other cell types did not express these genes (Figure 2). Using the Xenium platform with a 5,000-gene core and 100 custom genes, we spatially profiled 28 skin samples. Cell type annotations derived from scRNA-seq confirmed the presence and distribution of both pDC subsets. Neighborhood analysis revealed that pDC1s were proximity to B and T cells, whereas, conversely, pDC2s were localized near pDC1s and conventional dendritic cells (cDCs) (Figure 3).
Conclusion: This study provides new insights into early disease mechanisms in cutaneous lupus and identifies two transcriptionally distinct pDC populations associated with specific cellular niches and immune interactions in the skin, suggesting complex cellular hierarchy in driving inflammation and type I interferon responsesD in CLE.
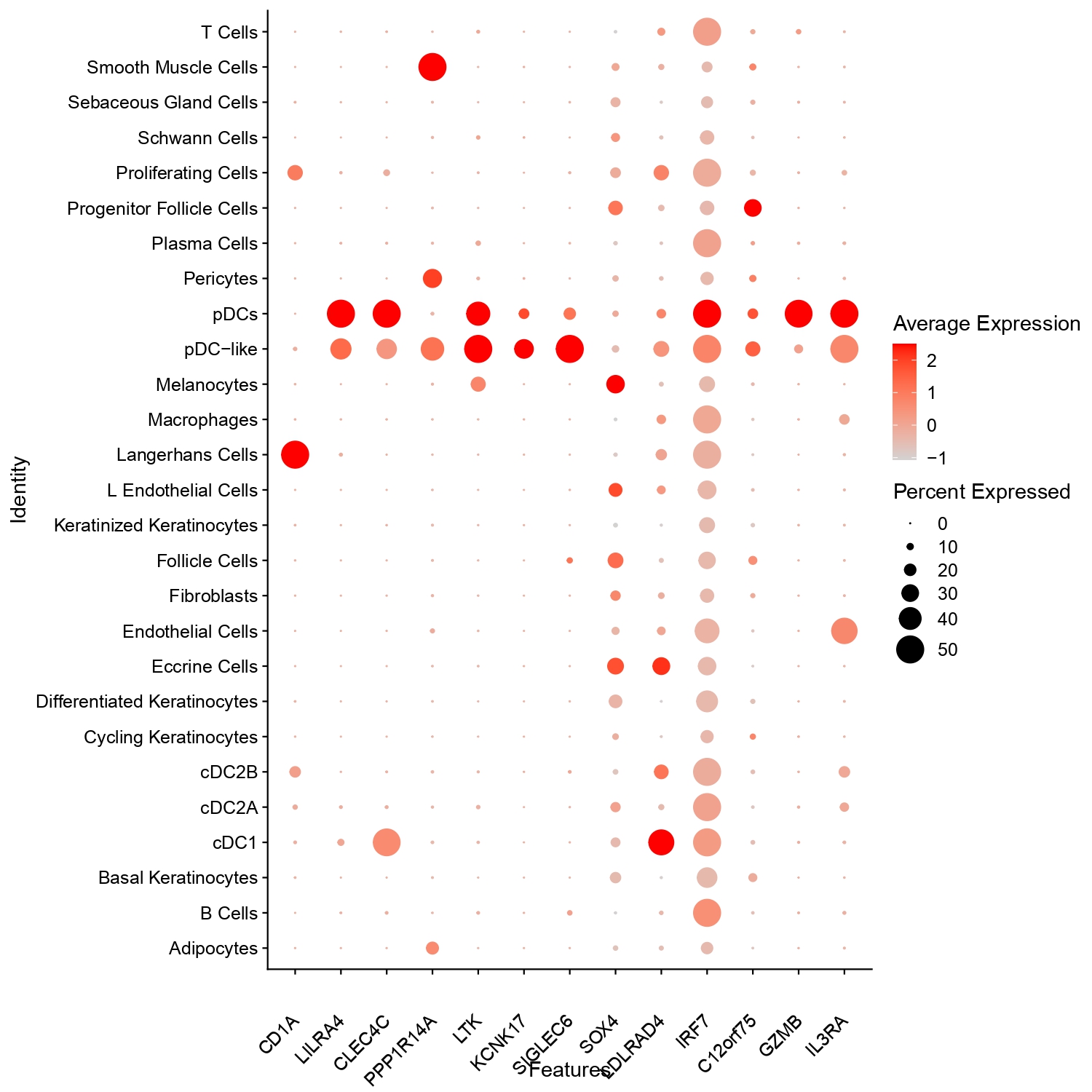 Figure 1: UMAP visualization of single-cell RNA-seq data in major- (A) and sub- (B) types. The distribution highlights transcriptionally distinct myeloid subsets and a pDC-like (pDC2) population relevant to autoimmune pathogenesis. Histogram illustrates proportions of each cell populations in different skin types (C).
Figure 1: UMAP visualization of single-cell RNA-seq data in major- (A) and sub- (B) types. The distribution highlights transcriptionally distinct myeloid subsets and a pDC-like (pDC2) population relevant to autoimmune pathogenesis. Histogram illustrates proportions of each cell populations in different skin types (C).
.jpg) Figure 2: Dot plot showing expression of selected genes across annotated skin and immune cell types. Dot size represents the percentage of cells expressing each gene, while color intensity reflects average expression. pDC and pDC-like populations show enriched expression of LILRA4, CLEC4C, IRF7, and GZMB, consistent with their transcriptional identity.
Figure 2: Dot plot showing expression of selected genes across annotated skin and immune cell types. Dot size represents the percentage of cells expressing each gene, while color intensity reflects average expression. pDC and pDC-like populations show enriched expression of LILRA4, CLEC4C, IRF7, and GZMB, consistent with their transcriptional identity.
.jpg) Figure 3: Spatial transcriptomic map of human skin biopsies (A). Cell identities of the spatial data were projected on scRNA-seq-derived annotations. Myeloid cell projection of the spatial data also confirms the pDC2 (pDC-like) population.
Figure 3: Spatial transcriptomic map of human skin biopsies (A). Cell identities of the spatial data were projected on scRNA-seq-derived annotations. Myeloid cell projection of the spatial data also confirms the pDC2 (pDC-like) population.
To cite this abstract in AMA style:
Werth V, Kang J, Lopes Almeida Gomes L, Richardson C, Chong B, Kahlenberg J, Guthridge J, DeJager W, Macwana S, Marlin C, James J, Bogle R, Tsoi A, Gudjonsson J. Cellular Landscape of Cutaneous Lupus Erythematosus Revealed by Single-Cell RNA Sequencing and Spatial Transcriptomics in the Lupus Accelerating Medicine Partnership Cohort [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/cellular-landscape-of-cutaneous-lupus-erythematosus-revealed-by-single-cell-rna-sequencing-and-spatial-transcriptomics-in-the-lupus-accelerating-medicine-partnership-cohort/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/cellular-landscape-of-cutaneous-lupus-erythematosus-revealed-by-single-cell-rna-sequencing-and-spatial-transcriptomics-in-the-lupus-accelerating-medicine-partnership-cohort/
