Session Information
Session Type: Abstract Submissions (ACR)
Background/Purpose: Rheumatoid arthritis (RA) is a heterogeneous disease. Predictive biomarkers have the potential to enhance treatment effectiveness by enabling identification of likely responders to a specific therapy.1,2 Our previous analysis of RA synovium showed the existence of 3 large subsets characterized by gene expression signatures suggestive of lymphoid, myeloid, or fibroid biology.3 In the present study, we demonstrated the ability of serum proteins produced by genes selected from those signatures to recapitulate the segregation of patients into these subsets.
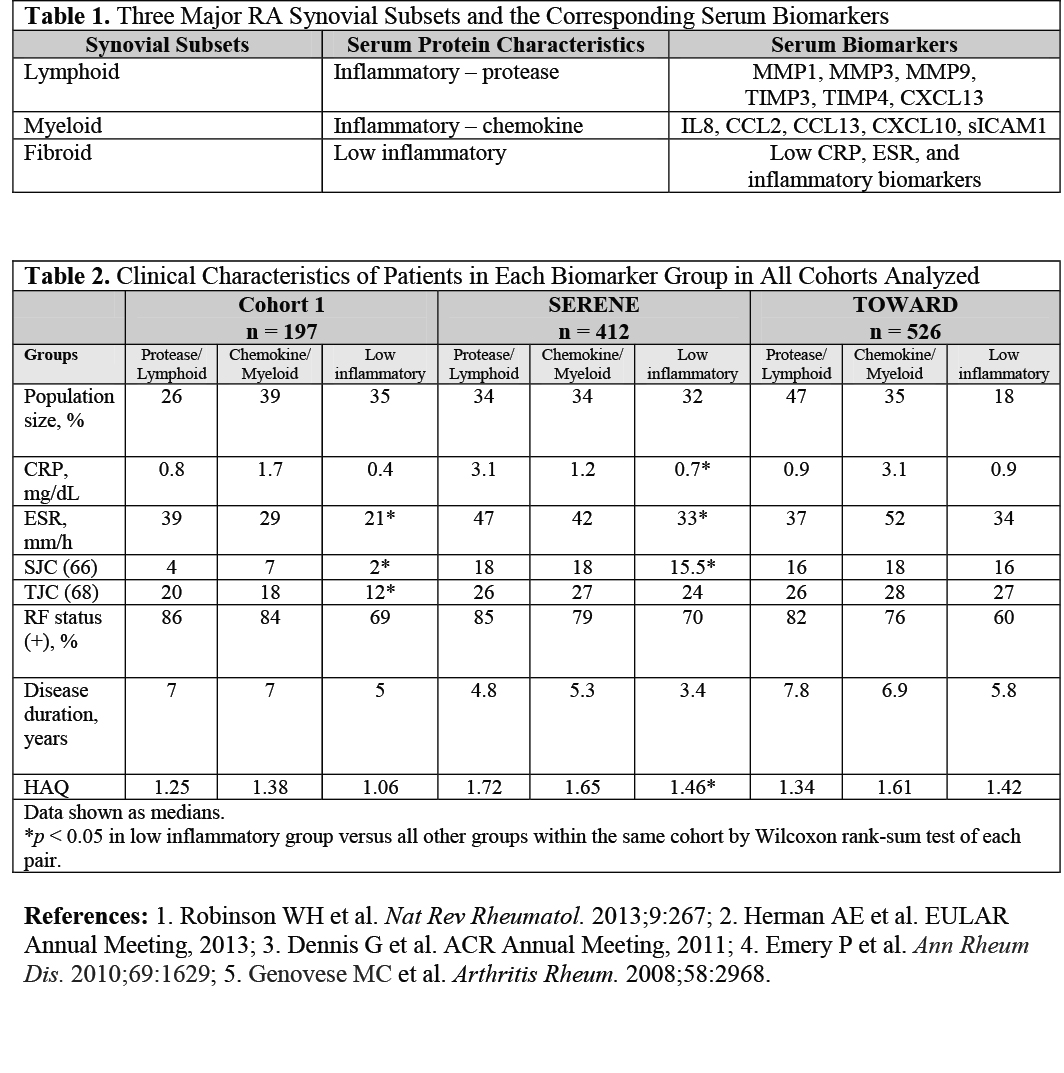
Methods: Several protein biomarker candidates were selected based on differential gene expression in 1 RA synovial subset versus the others and on detectability in serum. Quantitative immunoassay methods were used to measure these proteins in baseline serum from 3 biologic-naive RA cohorts: Cohort 1 (cross-sectional natural history cohort, n = 197), SERENE4 (n = 412), and TOWARD5 (n = 526). Levels of serum proteins were also compared between Cohort 1 patients and healthy controls (n = 30). Serum protein data were used to cluster patients and proteins. Patients were clustered into a specified number of groups using Partitioning Around Medoids (PAM), whereas proteins were clustered hierarchically. Each clinical characteristic of patients in each biomarker group was compared to the respective characteristic in all other groups within the same cohort using the Wilcoxon rank-sum test of each pair.
Results: Analysis of baseline serum biomarker levels in Cohort 1 showed that MMP1, MMP3, MMP9, TIMP3, TIMP4, CXCL13, IL8, CCL2, CCL13, CXCL10, and sICAM1 were elevated compared to those in healthy controls (p < 0.0001). These analytes had a wide dynamic range in all RA cohorts analyzed. The patient populations within each RA cohort segregated reproducibly into 3 groups, named by the characteristics of the serum proteins in each group: Protease, Chemokine, and Low Inflammatory. The 2 inflammatory groups (Protease and Chemokine) were dominated by serum biomarkers related to lymphoid and myeloid biology, respectively (Table 1). The third group generally displayed lower levels of inflammatory proteins, CRP, and ESR and was less often rheumatoid factor positive. This group likely corresponded to the synovial fibroid subset. The clinical characteristics of each biomarker group are summarized in Table 2.
Conclusion: We have identified serum biomarkers that segregate groups of patients associated with lymphoid and myeloid subtype–derived biomarkers in 3 large RA cohorts. These subpopulations of RA patients with distinct pathobiology can possibly demonstrate differential clinical response to treatments that target different inflammatory mediators. In addition, a low-inflammatory group was observed in all cohorts analyzed. This group is less likely to respond well to current biologic anti-inflammatory therapy.
Disclosure:
A. F. Setiadi,
Roche Pharmaceuticals,
1,
Roche Pharmaceuticals,
3;
N. Lewin-Koh,
Genentech and Biogen IDEC Inc.,
3;
S. Kummerfeld,
Genentech and Biogen IDEC Inc.,
3;
D. F. Choy,
Genentech and Biogen IDEC Inc.,
3;
C. T. J. Holweg,
Genentech and Biogen IDEC Inc.,
3;
S. Fischer,
Genentech and Biogen IDEC Inc.,
3;
A. Song,
Genentech and Biogen IDEC Inc.,
3;
M. J. Townsend,
Roche Pharmaceuticals,
1,
Genentech and Biogen IDEC Inc.,
3.
« Back to 2013 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/synovial-subset-derived-baseline-serum-biomarkers-segregate-rheumatoid-arthritis-patients-into-subgroups-with-distinct-serum-protein-and-clinical-characteristics/