Session Information
Date: Tuesday, November 14, 2023
Title: (1827–1839) Fibromyalgia & Other Clinical Pain Syndromes Poster
Session Type: Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Fibromyalgia (FM) is characterized by widespread pain, non-restorative sleep, fatigue, and cognitive dysfunction. TNX-102 SL is a sublingual cyclobenzaprine tablet designed for daily use at bedtime. SNPs from participants in two Phase 3 trials were analyzed by an iterative genome-wide association study (GWAS) method to build a set of drug-response-related alleles that correlate with improvements in pain and sleep.
Methods: Whole exome sequencing (WES) was performed on peripheral blood from participants in two Phase 3 trials of TNX-102 SL 5.6 mg in FM. Markers were identified using a multi-loci mixed model (MLMM) extended to include a genome x treatment arm (G×T) term. A GWAS was run to test for each marker. First, a model was made using only the top marker. The GWAS was then repeated using the first marker as a co-variate to find the next top marker for the model. This procedure was iterated until an optimal model was constructed and a panel of best-performing loci was identified. A scoring system was developed that counted the number of favorable alleles (0-36) present in each patient. We examined the correlation of this count score with drug response on pain and sleep across both studies.
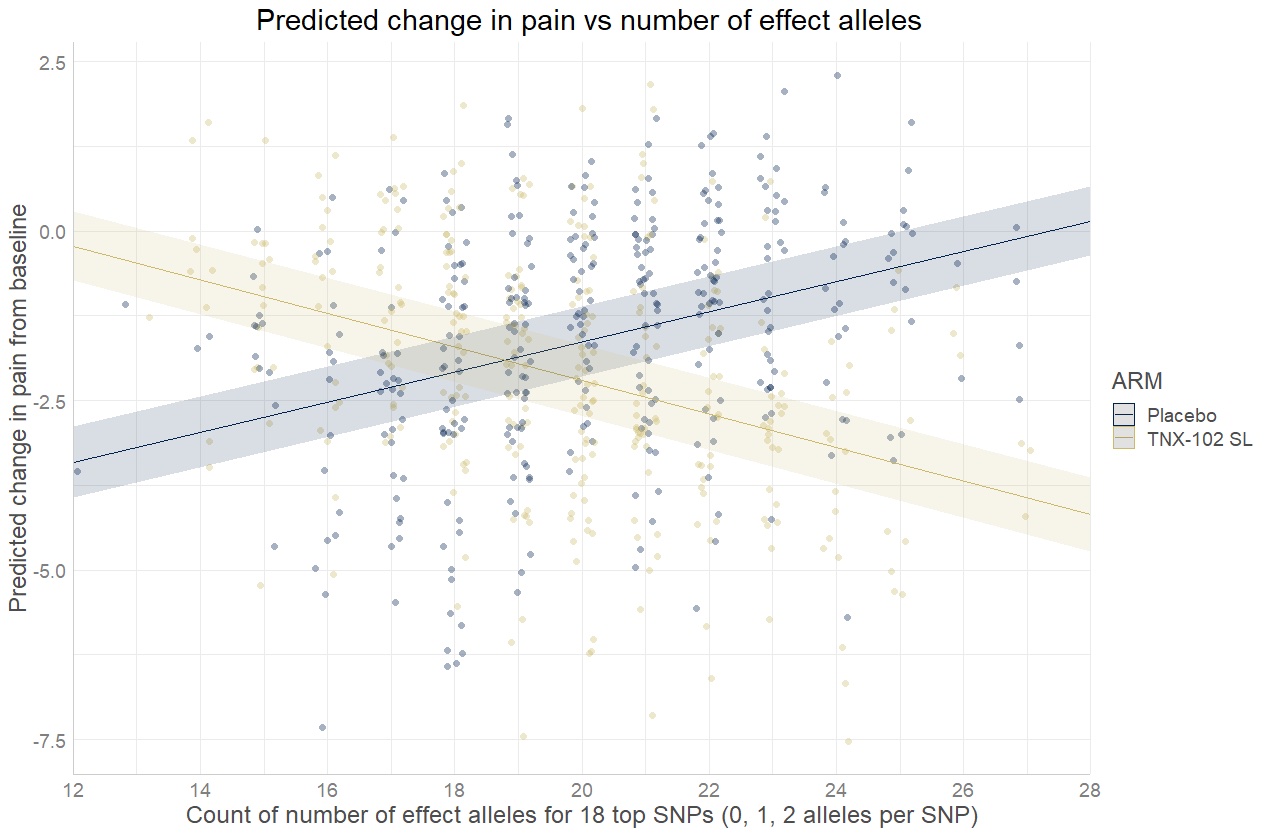
Results: In analyzing treatment responses in two Phase 3 FM studies, we identified 18 loci of interest. All of the markers in the final model had p-values < e-4 which are individually of interest, but not genome-wide significant. However, when used together to generate a count score, substantially improved predictive power was observed. Sub-setting patients based on the count score resulted in increased effect sizes for the change in pain score and sleep score when comparing TNX-102 SL to placebo in each study separately. As the count scores increase, the effect size improves while the sample size decreases. For example, if patients with at least a count score of 20 are retrospectively grouped, the effect size for pain reduction in the group increases to 0.90, as compared to 0.26 in the unselected population. Of note, all of the drug-response-related SNPs were intronic.
Conclusion: Polygenic drug-response related SNP-counts have potential to identify patients with higher effect size on pain reduction from TNX-102 SL treatment compared to placebo, as first explored in a retrospective analysis of two Phase 3 FM studies. The use of this polygenic SNP-counting method has the potential to identify patients in future studies that are likely to benefit from TNX-102 SL treatment. The Whole Genome Sequencing of study participants has been completed and may reveal additional drug-response-related SNPs. The panel will be validated on a future trial of similar design.
To cite this abstract in AMA style:
Rosenfeld J, Linse G, Slipher S, Flint C, Iserson A, Harris H, Engels J, Sullivan G, Lederman S. Development of a Polygenic Risk Model for Therapeutic Response to Bedtime Sublingual Cyclobenzaprine (TNX-102 SL*) in Fibromyalgia Based on Polygenic Single Nucleotide Polymorphism (SNP)-Count Scores [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/development-of-a-polygenic-risk-model-for-therapeutic-response-to-bedtime-sublingual-cyclobenzaprine-tnx-102-sl-in-fibromyalgia-based-on-polygenic-single-nucleotide-polymorphism-snp-count-scores/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/development-of-a-polygenic-risk-model-for-therapeutic-response-to-bedtime-sublingual-cyclobenzaprine-tnx-102-sl-in-fibromyalgia-based-on-polygenic-single-nucleotide-polymorphism-snp-count-scores/