Session Information
Session Type: Poster Session C
Session Time: 1:00PM-3:00PM
Background/Purpose: Overexpression of IL-23 using IL-23 minicircle DNA in B10.RIII mice results in the development of a spondyloarthritis-like disease. B10.RIII (B10.RIII-H2r H2-T18b/(71NS)SnJ) is a major histocompatibility complex (H-2) congenic strain derived from a cross between the donor strain RIII/WySn (RIII, H-2r) and the background strain C57BL/10Sn (B10, H-2b), followed by seven backcrosses to the background strain. In contrast to B10.RIII mice, B10 mice do not develop IL-23 minicircle-induced arthritis. This difference in phenotype is accounted for by the differences in the genetic backgrounds of the two strains. We set out to identify genomic regions determining arthritis susceptibility in this model.
Methods: The genome sequences of B10.RIII and B10 mice (B10.RIII(71NS)/Sn, C57BL/10SnJ, C57BL/10J) were compared using data from the European Variation Archive. Subcongenic mice were generated replacing individual RIII-derived regions in the B10.RIII genome (r allele) with the B10 interval (b allele). Adult male mice were hydrodynamically injected with 50 ng IL-23 EEV (System Biosciences), and arthritis development was monitored every other day for two weeks. Splenocytes from B10.RIII and B10 mice were sorted into 8 immune cell subsets and analyzed by bulk RNA-seq.
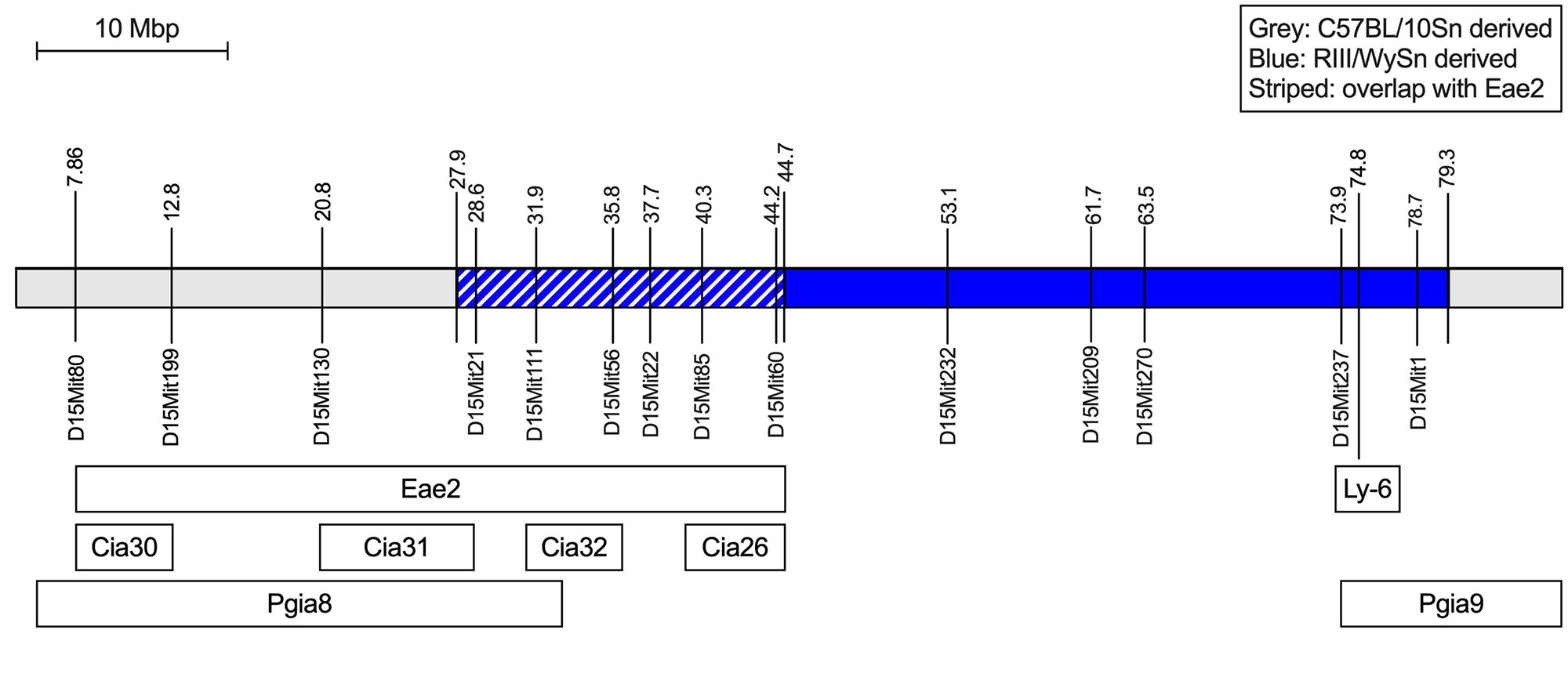
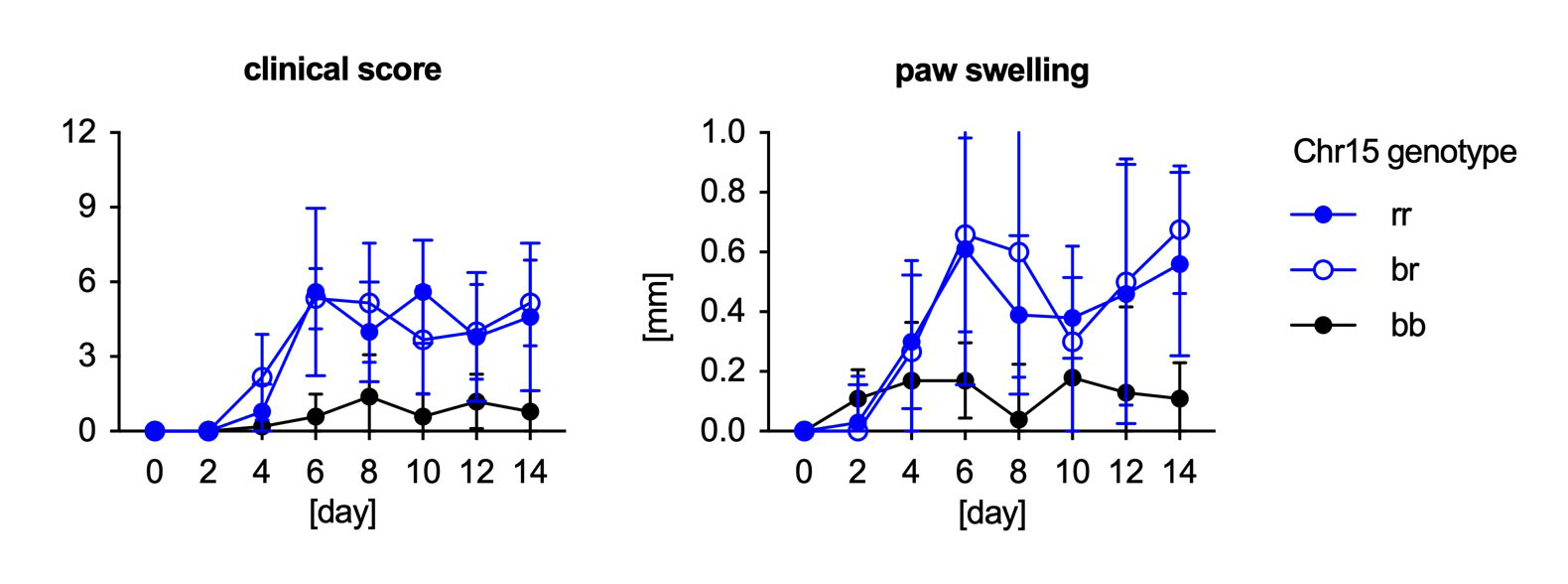
Results: Genome sequence analysis of B10.RIII and B10 strains confirmed RIII-derived regions in the B10.RIII chromosomes 10 (52 Mbp, previously reported) and 17 (25 Mbp, containing the congenic H-2r region). In addition, a small telomeric region on chromosome 14 (5 Mbp) and a large region on chromosome 15 (57 Mbp) were identified as RIII-derived. The RIII-derived regions in B10.RIII contain variants predicted to be damaging to the gene function and these variants are absent in the parental B10 strains. The RIII-derived region on chromosome 15 overlaps with arthritis QTLs previously identified in other models (Figure 1). Experiments with subcongenic strains showed that the RIII-derived regions on chromosomes 15 and 17 but not the chromosome 10 region affect susceptibility to IL-23 minicircle-induced arthritis in an autosomal dominant fashion (Figure 2). RIII-derived clusters accounted for a higher proportion of the differentially expressed genes in immune cell subsets from B10.RIII and B10 mice. Several genes carrying damaging variants were differentially expressed. We integrated genome sequence variation and gene expression data with information on gene function, arthritis QTLs, and arthritis-associated genes in humans to identify candidate genes for further analysis.
Conclusion: Susceptibility of B10.RIII mice to IL-23 induced spondyloarthritis-like disease is controlled by two major RIII-derived genomic regions, the congenic H-2r on chromosome 17 and a “contaminating” region on chromosome 15 that overlaps with arthritis susceptibility loci previously identified in other models. Candidate susceptibility genes include the Ly6 cluster on chromosome 15 and Ilrun on chromosome 17.
To cite this abstract in AMA style:
Soundararajan J, Shivalikanjli A, Keane T, Ermann J. Mapping Spondyloarthritis Susceptibility Loci in B10.RIII Mice [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/mapping-spondyloarthritis-susceptibility-loci-in-b10-riii-mice/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/mapping-spondyloarthritis-susceptibility-loci-in-b10-riii-mice/