Session Information
Date: Saturday, November 12, 2022
Title: Vasculitis – Non-ANCA-Associated and Related Disorders Poster I: Giant Cell Arteritis
Session Type: Poster Session A
Session Time: 1:00PM-3:00PM
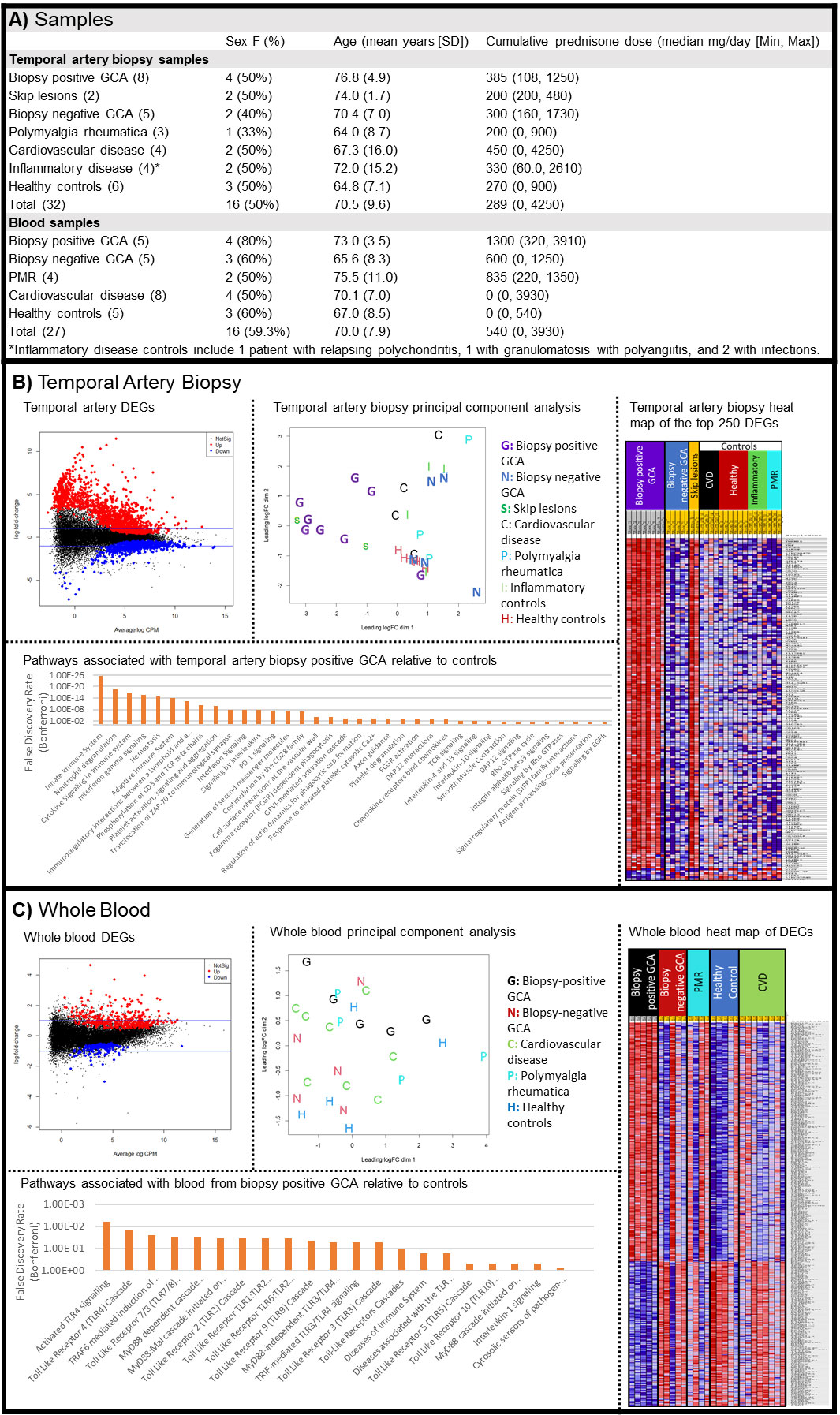
Background/Purpose: Diagnosing giant cell arteritis GCA is notoriously challenging, as there are no reliable blood biomarkers and temporal artery biopsy (TAB) has a sensitivity of only approximately 80%. Thus, patients suffer both from missed diagnoses and over-treatment. This study seeks to identify differentially expressed genes (DEGs) in the blood and TAB of patients with GCA relative to controls, as determined by RNA-sequencing (RNA-Seq).
Methods: Blood (PAXgene tubes) and formalin-fixed paraffin-embedded TAB tissue from patients undergoing a workup for suspected GCA were evaluated by RNA-Seq. Samples representative of biopsy positive GCA, biopsy negative GCA (negative biopsy but clinically diagnosed with GCA), polymyalgia rheumatica (PMR), non-vasculitic ischemia, other inflammatory diseases, and controls (negative TAB and not diagnosed with any disease) were included. Significant DEGs had a fold change (FC) > 1.5 and a false discovery rate (FDR) < 0.05. Pathway analysis of significantly DEGs was performed using Reactome. Age, sex, and cumulative corticosteroid exposure pre-sample collection were included as covariates in the models.
Results: 32 TAB and 27 whole blood samples were included in the analysis, including 8 biopsy positive GCA TABs and 5 biopsy positive GCA whole blood samples. (Fig. 1A) The majority of patients had glucocorticoid exposure pre-sample collection. TABs (Fig. 1B) of biopsy positive GCA revealed 1493 up- and 681 down-regulated transcripts relative to control groups (non-vasculitic ischemia, PMR, other inflammatory diseases, and controls). Skip lesions samples (n=2), which were dissected away from positive lesions, overlapped with biopsy positive GCA. However, biopsy negative GCA TABs showed no DEGs relative to controls. Pathways associated with biopsy positive GCA expression included innate immune pathways, interferon gamma and other cytokine signalling, PD-1, and integrin signalling. Whole blood (Fig. 1C) of biopsy positive GCA revealed 351 up- and 168 down-regulated transcripts relative to controls (healthy controls, cardiovascular disease, and PMR), however there was overlap with control groups. Pathways associated with biopsy positive GCA DEGs included toll like receptor pathways. Biopsy negative GCA blood did not show any significant DEGs relative to controls.
Conclusion: The TAB gene expression signature distinguishes biopsy positive GCA from controls. A preliminary analysis of skip lesions shows substantial overlap with biopsy positive GCA, which suggests that skipped areas may represent early lesions expressing molecular markers of disease. The lack of similarly between skip lesions and biopsy negative GCA tissue suggests that skip lesions are not a common cause of biopsy negative GCA. Whole blood from biopsy positive GCA gene shows a small number of DEGs and an overlap with controls. Whole blood of biopsy negative GCA is not substantially different from controls. Strengths include the use of common GCA mimics as controls and the evaluation of both biopsy positive and biopsy negative GCA. Limitations include a small samples size and the difficulty in accurately diagnosing biopsy negative GCA.
To cite this abstract in AMA style:
Friedman M, Albert D, Choi D, Stiefel H, Oglesbee-Venghaus C, Ogle K, Harrington C, Wilson D, Rosenbaum J. Gene Expression Profile of Temporal Artery Tissue and Whole Blood in Giant Cell Arteritis [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/gene-expression-profile-of-temporal-artery-tissue-and-whole-blood-in-giant-cell-arteritis/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/gene-expression-profile-of-temporal-artery-tissue-and-whole-blood-in-giant-cell-arteritis/