Session Information
Date: Saturday, November 12, 2022
Title: SLE – Diagnosis, Manifestations, and Outcomes Poster I: Diagnosis
Session Type: Poster Session A
Session Time: 1:00PM-3:00PM
Background/Purpose: It is known that the expression of type I Interferon (IFN) genes is upregulated in systemic lupus erythematosus (SLE). In this study, we determined an IFN gene signature by calculating a score from the expression of four IFN stimulated genes (IFI27, IFI44L, IFIT1, RSAD2). We evaluated the stability of the IFN4 signature over time in cohorts of SLE patients versus healthy donors and in patients with rheumatoid arthritis (RA) or fibromyalgia (FMS).
Methods: Results were obtained with a prototype assay* on the BioFire® system using PAXgene blood RNA tubes without undergoing RNA isolation. After RNA isolation from the same tubes, the IFN gene signature was measured through amplification-free digital detection using the nCounter® technology (NanoString, Seattle, WA). Normalized expression values were calculated as follows. For the BioFire system, the expression of each gene was calculated by 2^-ΔCT, the geometric mean of 3 endogenous controls was used for normalization (Mommert et al., 2021; Pescarmona et al., 2019). For the nCounter technology, the count number of the four IFN genes was normalized by the geometric mean of the count number of 16 endogenous controls, as well as the positive and negative controls with the nSolver software (Pescarmona et al., 2019). With both techniques, the relative gene expression of each interferon response gene was calculated by dividing it by the median normalized expression of the respective IFN response gene from normal healthy donors. The IFN4 score is determined by the median of the relative gene expression of the four IFN stimulated genes.
*No regulatory review has been conducted; not an IVD test
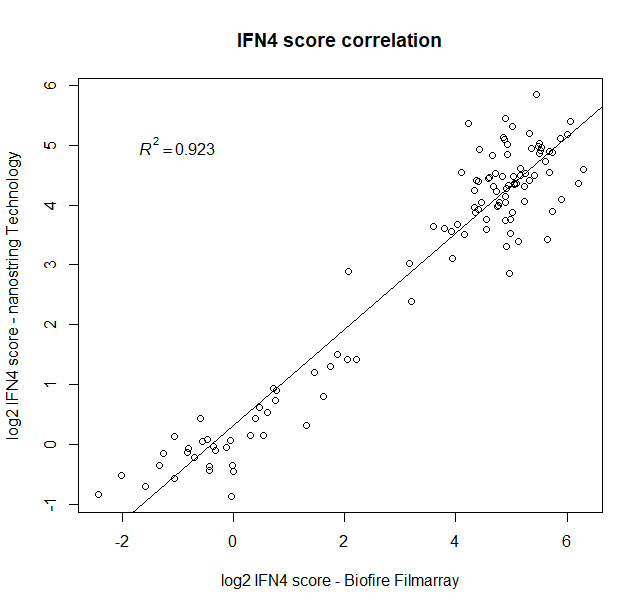
Results: Overall, a strong correlation between both platforms was observed when measuring expression of the four IFN signature genes in two consecutive visits in 47 established SLE patients (male [M]=4, female [F]=43), and 20 normal healthy donors (M=9, F=11) (Fig1).
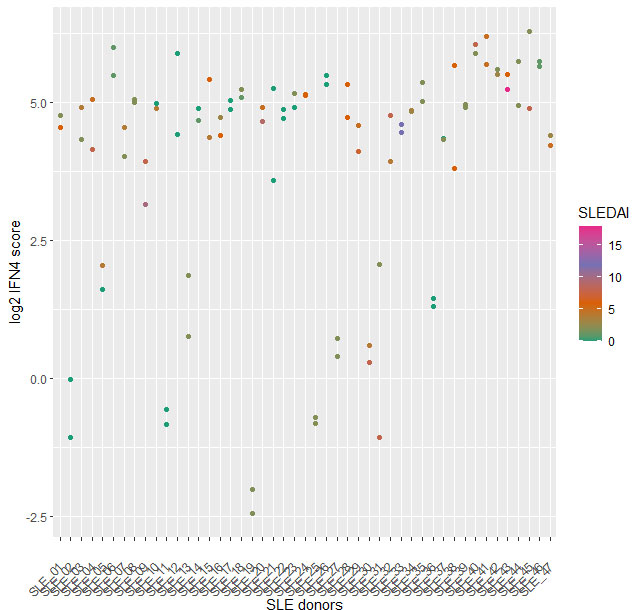
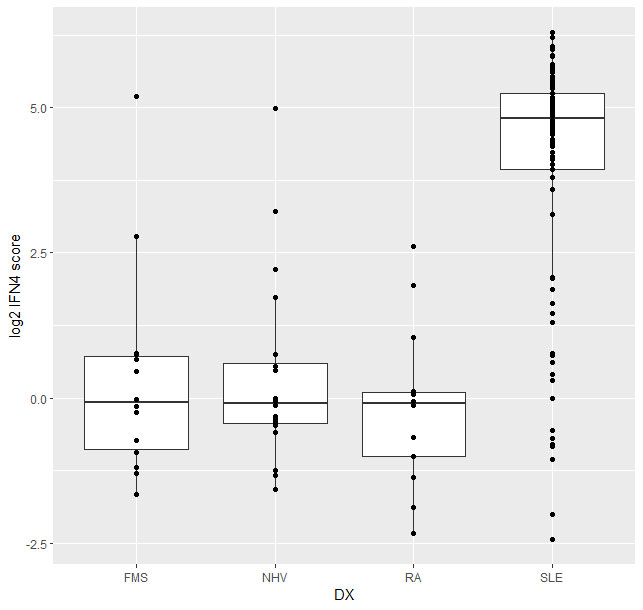
The IFN4 gene signature proved to be relatively stable in SLE over multiple visits and did not systematically track with disease severity (Fig2) or with disease subtypes. Analysis of 14 subjects with RA and 14 with FMS (all female 21% ANA+ in both groups) with the prototype assay on the BioFire system suggested that subjects with either disease exhibit a significantly lower IFN4 gene signature compared to SLE patients (Fig3).
Conclusion: In conclusion, we evaluated the IFN4 gene signature in SLE, RA and FMS by measuring the expression of four IFN stimulated genes (IFI27, IFI44L, IFIT1, RSAD2) with two different technologies. We demonstrate that the IFN4 signature in subjects with SLE is significantly elevated compared to subjects with RA or FMS. Furthermore, the IFN4 gene signature does not correlate with disease activity in SLE. Since the data between the two platforms correlate well with each other, both technologies could be effectively leveraged for small gene signatures analysis.
To cite this abstract in AMA style:
Kolb L, Mommert M, Brengel-Pesce K, Alexander R, Kyttaris V, Kammesheidt A, Stephens G. Comparison of NCounter® and BioFire® Technologies for the Measurement of Type I Interferon Signature [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/comparison-of-ncounter-and-biofire-technologies-for-the-measurement-of-type-i-interferon-signature/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/comparison-of-ncounter-and-biofire-technologies-for-the-measurement-of-type-i-interferon-signature/