Session Information
Date: Monday, November 8, 2021
Session Type: Poster Session C
Session Time: 8:30AM-10:30AM
Background/Purpose: To determine whether an eventual diagnosis of giant cell arteritis in both temporal artery biopsy positive and negative patients can be identified using prospective clinical factors, either through direct identification or machine learning techniques.
Methods: All patients at a single center who underwent temporal artery biopsy between January 2011 and November 2020 with accessible progress notes were included. Final diagnosis was determined from the documented clinical disease course after a minimum of one year, and was ascertained from the medical record by two assessors, with arbitration by a third assessor for cases of disagreement. Models were trained to distinguish GCA from mimics, with further comparisons performed on the subgroups of biopsy-positive and biopsy-negative GCA. Variables considered included pathology, clinical and demographic features (Table 1). Missing variables were imputed using k-nearest neighbours and the minority class was upsampled using SMOTE(1). After hyperparameter tuning, random forest models were fitted to the data(2), using 50-repeated 5-fold cross-validation to determine the out-of-sample area under the receiver operating characteristic (AUC). To assess individual variable contribution to model predictions, Shapley scores were calculated and compared between the three groups(3).
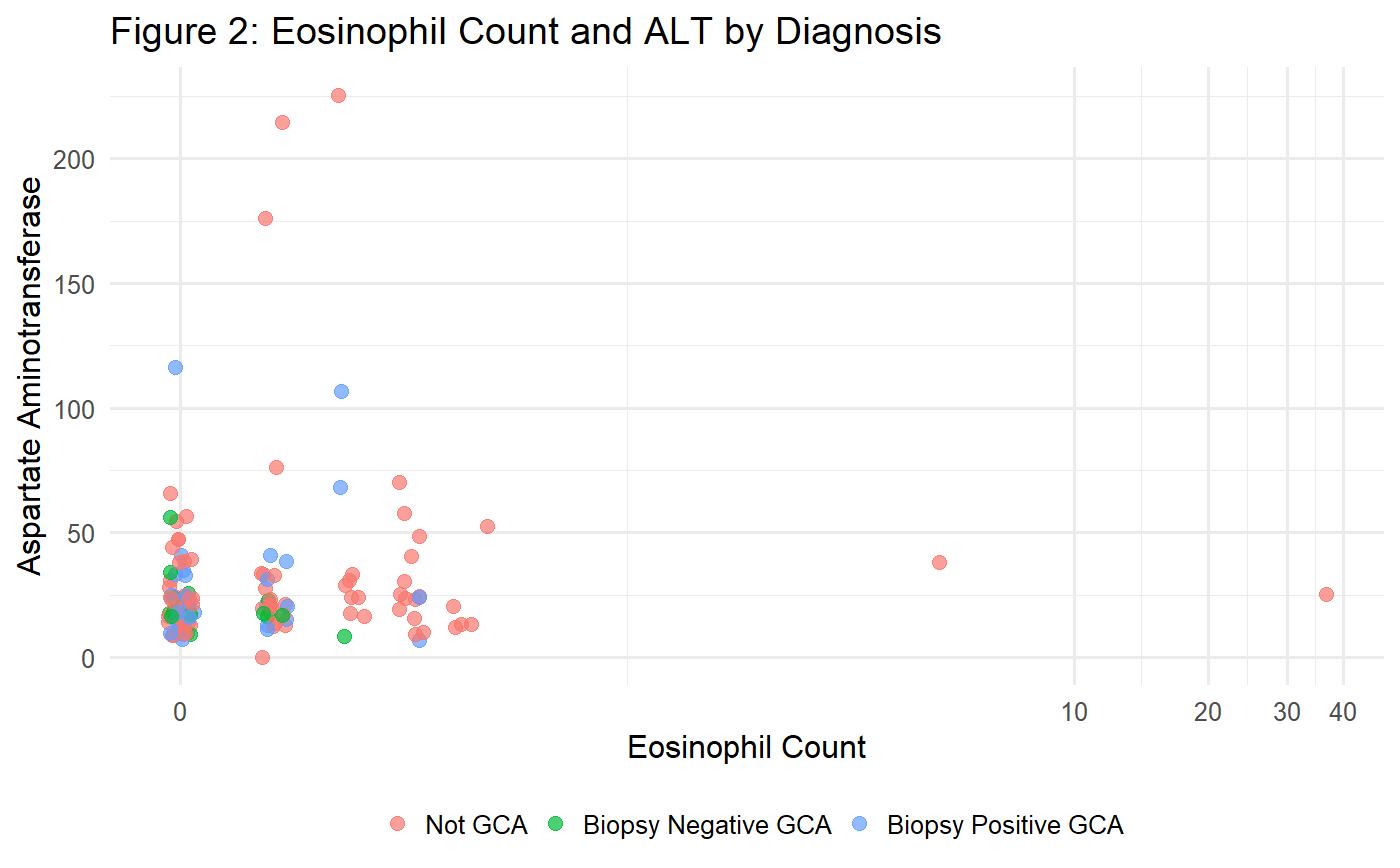
Results: During the study period, 194 patients underwent temporal artery biopsy (130 not GCA, 19 biopsy-negative GCA, 45 biopsy-positive GCA). The mean AUC of the random forest classifier was 0.726 (95% CI 0.715 – 0.737), with an overall classification accuracy of 70.4%. The Shapley scores demonstrate that, whilst the platelet count strongly predicts biopsy-positive GCA, the model must rely on a variety of other variables to predict biopsy-negative GCA. In particular, the eosinophil count, patient age, and liver function tests were discriminators of biopsy-negative GCA (Figure 1). The presence of an elevated eosinophil count or elevated aspartate aminotransferase distinguishes GCA mimics from biopsy-negative GCA (Figure 2).
Conclusion: Platelets, eosinophils, age, and ALT assist in differentiating biopsy-negative GCA, but discriminating GCA from mimics is best performed by machine learning models. In particular, the more difficult cases of biopsy-negative GCA may be distinguishable using models that use both positive markers of GCA diagnosis and variables that predict GCA mimics. Predictive models should therefore be developed on datasets that consider both predictors of GCA and its mimics.
- Hvitfeldt E. themis: Extra Recipes Steps for Dealing with Unbalanced Data [Internet]. 2020. Available from: https://CRAN.R-project.org/package=themis
- Wright MN, Ziegler A. ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. Vol. 77, Journal of Statistical Software. 2017. p. 1–17. Available from: http://dx.doi.org/10.18637/jss.v077.i01
- Greenwell B. fastshap: Fast Approximate Shapley Values [Internet]. 2020. Available from: https://CRAN.R-project.org/package=fastshap
Disclosures: C. McMaster, None; V. Yang, None; R. Buchanan, None; D. Liew, None.To cite this abstract in AMA style:
McMaster C, Yang V, Buchanan R, Liew D. Machine Learning Enhances the Identification of GCA from Its Mimics Based on Clinical Factors [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/machine-learning-enhances-the-identification-of-gca-from-its-mimics-based-on-clinical-factors/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/machine-learning-enhances-the-identification-of-gca-from-its-mimics-based-on-clinical-factors/