Session Information
Date: Sunday, November 7, 2021
Title: SLE – Diagnosis, Manifestations, & Outcomes Poster II: Manifestations (0855–0896)
Session Type: Poster Session B
Session Time: 8:30AM-10:30AM
Background/Purpose: Genetics and gene expression has been shown to correlate with systemic lupus erythematosus (SLE) disease severity. Our aim was to identify genetic risk loci for disease activity measures over time, representing disease activity burden.
Methods: The cohort study included participants from a tertiary care centre dedicated Lupus Clinic. Participants all met ≥4 of the ACR/SLICC classification criteria for SLE or 3 criteria and a positive biopsy. All participants had prospectively collected clinical data including repeated measures of disease activity evaluated with the Systemic Lupus Erythematosus Disease Activity Index 2000 (SLEDAI-2K). We restricted to participants with a first Lupus clinic visit within 3 years of SLE diagnosis and at least 2 clinic visits within 5 years. Participants were genotyped on the Illumina Multi-Ethnic Global Array, Global Screening Array or Omni 1-Quad array. Ungenotyped SNPs were imputed and ancestry was inferred using principal components (PCs) (1000 Genomes Project reference). The outcome was adjusted mean SLEDAI-2K and corticosteroids (AMSG), the calculated area under the curve of SLEDAI-2K over time, accounting for corticosteroid dose. Continuous square-root of AMS-G was the outcome of interest, transformed to improved normality. We included variants with minor allele frequency (MAF) of ≥0.01 and imputation quality r2 >0.3. Genome-wide additive allelic linear regression models were adjusted for sex and 5 PCs, stratified by array (MEGA+GSA and OMNI) and results were meta-analyzed using inverse variance weighting. A genome-wide significance threshold (p< 5x10-8) was used to indicate statistical significance.
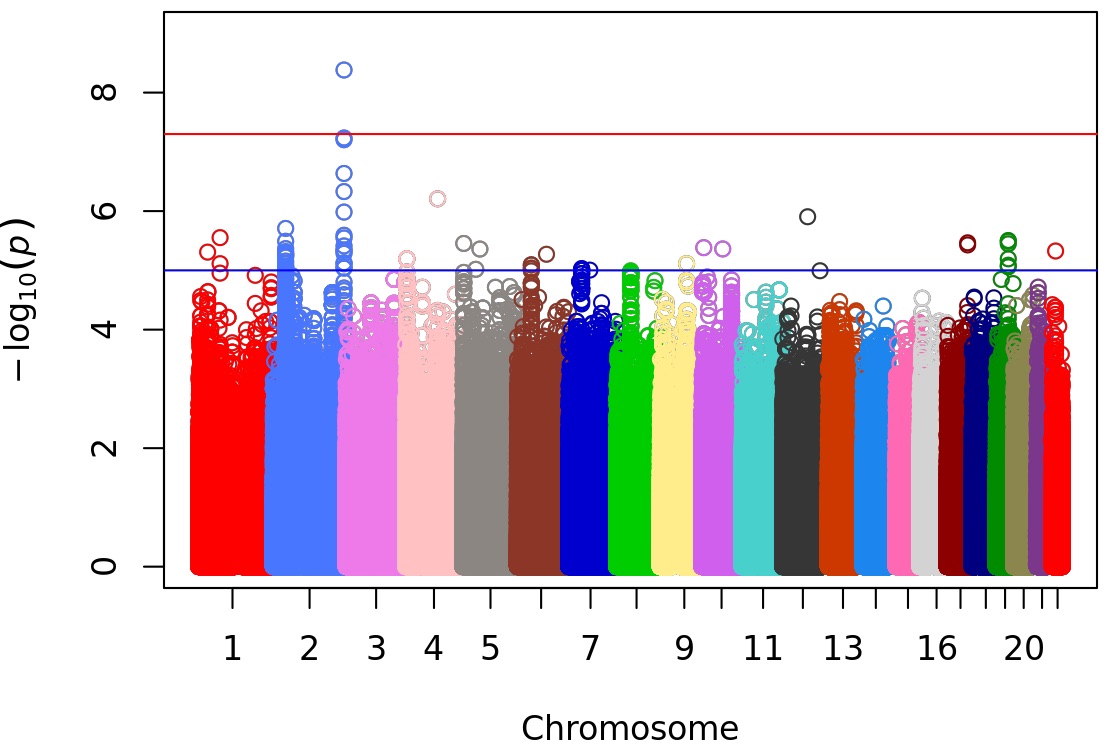
Results: The cohort included 538 individuals with SLE. The majority (75%) had a first clinic visit within 1 year of SLE diagnosis. The majority were of European ancestry (54%) with 16% of East Asian ancestry and 12% African ancestry. The median ASMG was 5.5 (IQR 3.2, 8.8). Meta-GWAS identified a genome-wide significant SNP for ASMG (rs4561613) on chromosome 2 intronic to AGAP1 (Beta 0.34, SE 0.06, P=4.2×10-9) (Figure). The overall MAF is 0.43, with a higher risk allele frequency for increased ASMG in non-European ancestral groups (0.67 in African ancestry; 0.41 East Asian ancestry) compared to those of European ancestry (0.36).
Conclusion: We identified a genome-wide significant locus for SLE disease activity burden as measured by AMSG. Variants in AGAP1 has been linked with body mass index and red blood cell indices. The mechanism by which AGAP1 variants impact disease activity and severity in SLE is unclear. Future studies are required to replicate these findings in independent cohorts.
To cite this abstract in AMA style:
Hiraki L, Liao F, Gladman D, Lee K, Touma Z, Wither J, Cook R, Urowitz M. Genetics of Longitudinal Disease Activity in Adults with Systemic Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/genetics-of-longitudinal-disease-activity-in-adults-with-systemic-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genetics-of-longitudinal-disease-activity-in-adults-with-systemic-lupus-erythematosus/