Session Information
Date: Monday, November 9, 2020
Title: Pediatric Rheumatology – Clinical Poster III: SLE, Vasculitis, & JDM
Session Type: Poster Session D
Session Time: 9:00AM-11:00AM
Background/Purpose: Genome wide association studies (GWAS) have identified >90 SNPs associated with systemic lupus erythematosus (SLE) risk. However, there may be additional loci impacting the age of diagnosis. The purpose of this study is to identify genetic variants for age of SLE diagnosis.
Methods: Our cohort included patients with childhood-onset SLE (cSLE) diagnosed < 18 years of age, and adult-onset SLE (aSLE), who met ACR and/or SLICC classification criteria for SLE. All patients were followed at tertiary care centers: Toronto Western Hospital, Hospital for Sick Children (Toronto), University of Manitoba (Winnipeg), Lurie Children’s Hospital of Chicago, St. Joseph's Health Centre (London, ON), and Hospital for Special Surgery (New York). We censored patients with age at diagnosis ≥ 70 years. Patients were genotyped on the Illumina Multiethnic Array (MEGA), and ungenotyped SNPs were imputed using the Haplotype Reference Consortium (HRC) reference. We restricted to SNPs with a minor allele frequency (MAF) ≥ 0.01 and imputation quality ≥ 0.8. Ancestry was genetically inferred from principal components (PCs) and ADMIXTURE calculated in reference to 1000 Genome Project (1KGP). Non-HLA, additive SLE weighted genetic risk scores (GRSs) were computed using published SLE GWAS log-odds ratio weights. Single-variant genome-wide linear regression of age of SLE diagnosis was performed with GENESIS. Multivariate models were adjusted for sex, aSLE/cSLE status, indicator for center, 5 PCs and SLE non-HLA GRS. We also completed a genome-wide test of cSLE risk (vs. aSLE) using a logistic regression model adjusted for the same covariates.
Results: Our cohort included 1093 patients, 88% female. 36% were of European ancestry, 23% East Asian and 18% Admixed. The median age at diagnosis was 17.1y (IQR 13.6, 30.8) (Table 1). We included 8.9M SNPs in GWAS. The most significant SNP associated with age at SLE diagnosis in the linear model was on chr11, rs138239231 (Beta 10.0y, SE 1.85y, P=5.73×10-8, MAF 0.01) upstream of DHCR7 and NADSYN1. DHCR7 encodes an enzyme involved in cholesterol metabolism, and NADSYN1 encodes a coenzyme involved in metabolic redox reactions and cell signalling. The second locus on chr14 (rs144180822: Beta 5.7y, SE 1.15y, P=7.15×10-7, MAF 0.03) is intronic to NUBPL, a gene encoding an iron/sulfur protein involved in the assembly of a mitochondrial inner membrane enzyme. In the logistic model, the most significant SNPs were on chr1, rs12024309 upstream to anti-sense RNA SMG7-AS1 (OR 0.5, [95% CI: 0.4, 0.7], P=1.16×10-6, MAF 0.39); chr4, rs10001705 upstream to HS3ST1, involved in the biosynthesis of anticoagulant heparin sulfate (OR 3, [95% CI:1.9, 4.6], P=1.31×10-6, MAF 0.11); on chr17, rs116981214, intronic to MRPL45P2, a mitochondrial ribosomal protein (OR 4.7, [95% CI: 2.5, 8.7], P=1.46×10-6, MAF 0.05). None of these loci reached genome-wide significance (P< 5x10-8).
Conclusion: In our multiethnic cSLE and aSLE cohort, GWAS did not identify a genome-wide significant SNP association with the age at diagnosis or cSLE risk. We identified 2 loci near genome-wide significance for age at SLE diagnosis, and 3 near genome-wide significance for cSLE risk. We plan to expand our analyses including more patients.
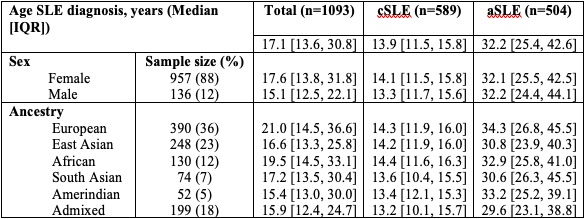 Table 1. Age at systemic lupus erythematosus diagnosis, overall and stratified by sex and ancestry
Table 1. Age at systemic lupus erythematosus diagnosis, overall and stratified by sex and ancestry
To cite this abstract in AMA style:
Carlomagno R, Liao F, Cao J, Gladman D, Klein-Gitelman M, Knight A, Levy D, Onel K, Paterson A, Peschken C, Pope J, Touma Z, Urowitz M, Webber D, Wither J, Silverman E, Hiraki L. Genetics of Age at Diagnosis in Systemic Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/genetics-of-age-at-diagnosis-in-systemic-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genetics-of-age-at-diagnosis-in-systemic-lupus-erythematosus/
