Session Information
Session Type: Late-Breaking Abstract Poster Session
Session Time: 9:00AM-11:00AM
Background/Purpose: Anti-citrullinated protein antibodies (ACPA) are currently the primary biomarker for identifying individuals at increased risk for future RA development. However, we have recently shown in a prospective study that most unaffected ACPA+ individuals do not develop RA. We hypothesized that abnormalities in the serum proteome may serve as additional biomarkers in the prediction risk for future disease onset. The aim of our study was to mine the serum proteome of individuals who ultimately developed RA to detect biomarkers that predict disease onset.
Methods: Using the SOMAscan (slow off-rate modified aptamer) array, SomaLogic, Boulder Co, we generated quantitative levels of 1307 proteins in serum samples from seventeen first-degree relatives (FDR) of Indigenous North American (INA) RA patients who developed inflammatory arthritis (IA) synovitis after having been followed prospectively for a mean of 3.2 years. All were ACPA+ at time of IA diagnosis. Each individual had two preclinical samples, one at the time of IA onset, and one at an earlier time point. We also analyzed samples from ACPA+ FDR (n = 63) and ACPA- FDR (n = 47) who did not develop inflammatory arthritis. Differentially expressed proteins were identified using ANOVA corrected for multiple comparisons. We applied a machine learning lasso regression model to identify a minimum set of proteins that classified patients who transition into clinical arthritis.
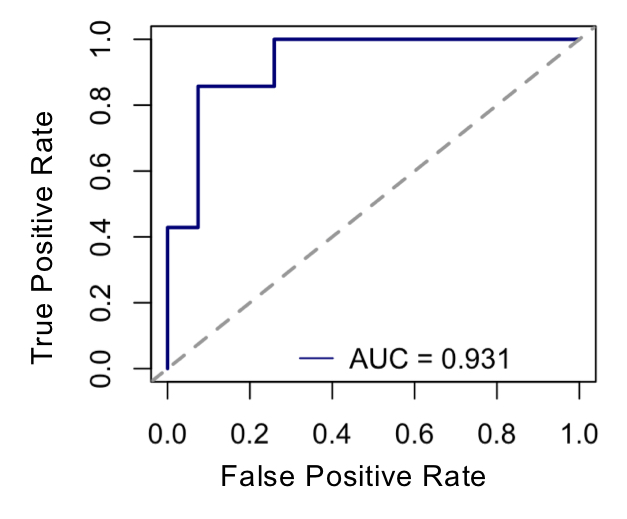
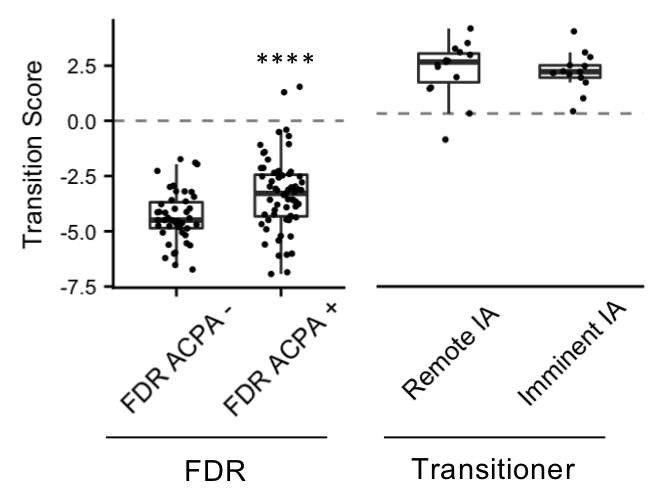
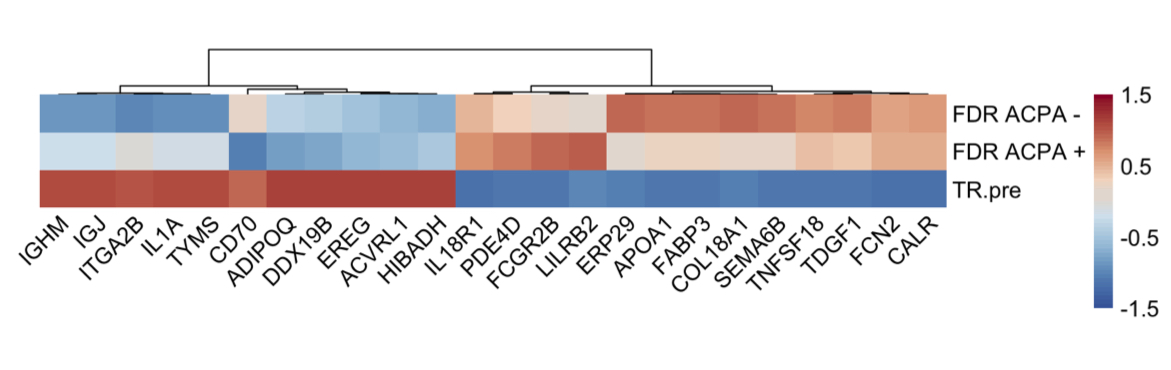
Results: Differential expression of 669 proteins (260 upregulated, 409 downregulated) were identified between pre-Transition samples (TR-pre) and ACPA negative FDR. ITGA2B and HIST1H3A were the highest upregulated proteins in TR-pre samples, while protease inhibitors SERPINA5 and ITIH4 were highly downregulated in the TR-pre samples. Compared with ACPA- FDR, overlap between TR-pre baseline and non-baseline samples of highly upregulated (log2 difference > 0.5) proteins was 60%, suggesting alterations in the serum proteome occur years before the development of RA. A lasso regression model identified a 23-marker panel that classified TR-pre samples from the larger pool of ACPA+ and ACPA- FDR serum samples. In a validation cohort (n = 34), the model correctly classified 31/34 samples (91.2% accuracy) with a sensitivity of 95.6% and specificity of 85.7%. Area under the curve (AUC) was 0.931 for the model. Transition scores were extracted from the model, which were higher in ACPA+ FDR compared to ACPA- FDR (p < 0.001). There were no differences in Transition score comparing TR-pre samples that were remote or close to the time of IA onset.
Conclusion: Compared to at-risk individuals who did not develop IA, clear and reproducible differences in the serum proteome are demonstrable in the serum samples of individuals who ultimately developed IA, even several years before the onset of clinically evident disease. Our findings suggest that a small serum biomarker panel can serve to accurately classify at-risk individuals who have a high likelihood of progressing to develop IA.
To cite this abstract in AMA style:
O'Neil L, Spicer V, Smolik I, Meng X, Wilkins J, El-Gabalawy H. A Serum Proteomic Signature Defines Transition from the Preclinical State to Rheumatoid Arthritis [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/a-serum-proteomic-signature-defines-transition-from-the-preclinical-state-to-rheumatoid-arthritis/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-serum-proteomic-signature-defines-transition-from-the-preclinical-state-to-rheumatoid-arthritis/