Session Information
Date: Monday, November 11, 2019
Title: B Cell Biology & Targets In Autoimmune & Inflammatory Disease Poster
Session Type: Poster Session (Monday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Dysregulation of B cells play a critical role in the pathogenesis of primary Sjögren’s syndrome (pSS). Long non-coding RNAs (lncRNAs), which have become the focus of studies on autoimmune diseases, may contribute to the pathogenesis of pSS through multiple signal transduction pathways. However, no study focuses on the effects of dysregulation of the transcriptomes, including lncRNAs, of B cells on the pathogenesis of pSS. The aim of this study was to identify the molecular mechanism of dysregulation of B cell subpopulations of pSS at the transcriptome level.
Methods: We enrolled patients with pSS (n=6) and healthy controls (HC) (n=6). Peripheral B cells acquired from these subjects were separated by cell sorting into four subsets: CD38–IgD+ (Bm1), CD38+IgD+ (naïve B cells), CD38highIgD+ (pre-germinal centre B cells), and CD38±IgD– (memory B cells). Total RNA was extracted, and gene expression was measured using microarrays. We conducted a bioinformatics analysis to identify differentially expressed genes (DEGs) and weighted gene co-expression network analysis (WGCNA) to identify pSS-associated signalling pathways. We validated gene expression levels in CD19+ B cells of patients with pSS (n=7) and HC (n=7) in other cohorts by quantitative PCR (qPCR).
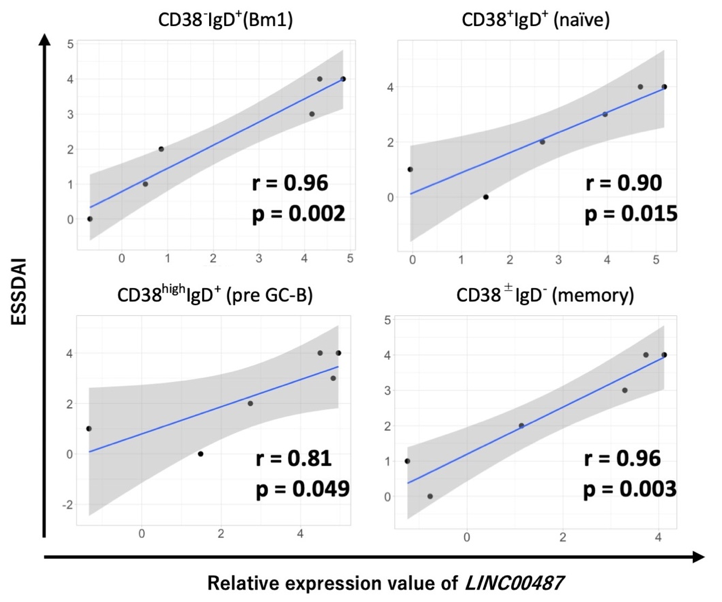
Results: Using principal component and clustering analyses, we found that transcript expression patterns depended on cell type rather than clinical condition (pSS or HC). DEGs analysis identified LINC00487 as significantly upregulated in all B cell subsets as well as HLA and interferon signature genes. Moreover, the normalized intensity value of LINC00487 significantly correlated with the disease activity score in all pSS B cell subsets (Figure 1). An in vitro study using human B cell lines revealed that the expression of LINC00487 was strongly induced by IFNα. In validation cohort using qPCR, expression of LINC00487 in CD19+ B cells of patients with pSS tended to upregulate compared with HC, although it was not significant (relative expression levels of LINC00487/GDH; 0.00227 vs 0.00025, p = 0.12). Further, expression of LINC00487 was significantly correlated with interferon stimulated gene, interferon induced protein 44 like (IFI44L). WGCNA revealed six clusters associated with the B cell subpopulation of pSS. Further, genes that encode components of the B cell receptor (BCR) signaling pathway were identified as intramodule hub genes such as IKZF3 and HRK. SOX4, which may be targeted by several microRNAs identified in upstream analysis, was identified as an inter-module hub gene (Figure 2).
Conclusion: Our transcriptome analysis revealed key genes involved in the molecular dysregulation of B cell subpopulations associated with pSS. This knowledge contributes to our understandings of the pathogenesis of pSS.
To cite this abstract in AMA style:
Inamo J, Suzuki K, Takeshita M, Kassai Y, Takigchi M, Kurisu R, Okuzono Y, Tasaki S, Takeuchi T. Identification of Novel Genes Associated with Dysregulation of B Cells in Patients with Primary Sjögren’s Syndrome [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/identification-of-novel-genes-associated-with-dysregulation-of-b-cells-in-patients-with-primary-sjogrens-syndrome/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-novel-genes-associated-with-dysregulation-of-b-cells-in-patients-with-primary-sjogrens-syndrome/