Session Information
Session Type: Poster Session (Sunday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Rapidly expanding collections of Electronic Medical Records (EMR) form a valuable resource for clinical research. Besides entries with a standardized format, EMRs often also contain free text fields intended for noting specifications of the treatment policy. While these free text fields contain essential information, their free nature makes them hard to parse, as they contain typos or acronyms. As a result, data extraction from EMR is often performed manually, or is performed while excluding the format-free fields. The purpose of this study is to develop and validate a text-mining approach to extract medication prescribed for Rheumatoid Arthritis as contained in format-free fields of an EMR.
Methods: The EMR dataset consisted of 45,012 entries from 2,771 patients that visited the rheumatology outpatient clinic from the Leiden University Medical Centre between 2007 and 2018. We randomly selected 15% and 7,5% of the entries to create a training and test set, with 5,992- and 2,993 entries respectively. The training set was used to design the algorithm, whereas the test set was used as an independent validation of the algorithm’s performance of identifying each of the DMARDs and biologicals routinely prescribed for treating RA.
Using methods derived from Natural Language Processing, we developed an algorithm that consecutively performs three tasks: 1. Text pre-formatting 2. Acronym recognition and 3. Typo correction. Text pre-formatting consisted of several simple operations to deal with the most prevalent textual artifacts, including separation of special characters and punctuation sticking to words. Ten independent clinicians compiled acronym lists for each of the routinely prescribed RA medication. Lastly, for typo correction, we employed the Damerau-Levenshtein1 (DL) distance that determines the similarity between two words by counting the number of single character operations (remove, add, move or replace) required to transform one word into another. Using the training set, we computed for each drug DL distances between all words in the free fields of the EMRs and a particular drug name or its acronym. Using the annotations created in the training set we then determined the DL distance optimally distinguishing between a typo and two similar words with a different meaning.
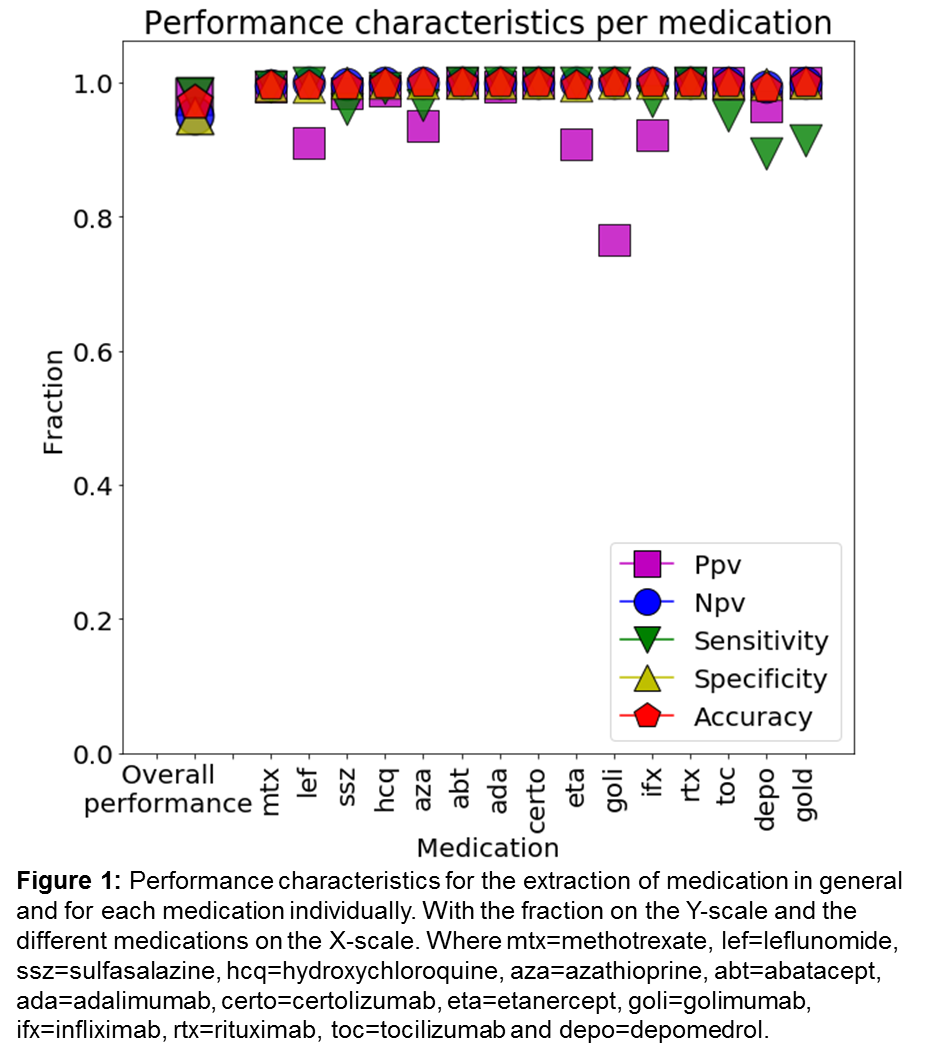
Results: Fifteen medications for the treatment of RA were present in our sample (see figure 1). In total, medication was present in 1,789 out of the 2,993 entries. The median DL cutoff for typos was 2 with a standard deviation of 0.96. The overall accuracy of our drug-identification-algorithm was very good per medication in general (0.97) and the individual test characteristics were high: sensitivity=0.98 and specificity=0.95, PPV=0.98, NPV=0.95. Also on an individual drug-level the performance was high: accuracy >=0.99, sensitivity >=0.89 and specificity >=0.99, NPV >=0.99 and PPV >=0.90 for all medication except golimumab.
Conclusion: We developed and validated an algorithm enabling a highly accurate automated extraction of RA medication from format-free fields of Electronic Medical Records.
To cite this abstract in AMA style:
Maarseveen T, Huizinga T, Reinders M, van den Akker E, Knevel R. A Validated Text-mining Algorithm to Extract Rheumatoid Arthritis Medication Contained in Format-free Fields of Electronic Medical Records [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/a-validated-text-mining-algorithm-to-extract-rheumatoid-arthritis-medication-contained-in-format-free-fields-of-electronic-medical-records/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-validated-text-mining-algorithm-to-extract-rheumatoid-arthritis-medication-contained-in-format-free-fields-of-electronic-medical-records/