Session Information
Date: Tuesday, October 23, 2018
Title: Systemic Lupus Erythematosus – Etiology and Pathogenesis Poster III
Session Type: ACR Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Systemic lupus erythematosus (SLE) is a multifactorial disease with genetic and environmental risk factors, and heterogeneous manifestations that encompass a wide range of disease severity. Long-term outcomes for individual patients are difficult to predict. Little is known about why an affected individual might develop a particular SLE phenotype. Previous studies have used phenotype-mapping approaches to identify subtypes of SLE using genome-wide association studies and gene expression data; however, studies integrating both genetic and clinical data to identify SLE phenotypes using bioinformatics analyses remain limited.
Methods: We characterized subgroups of patients using sociodemographic, clinical and genetic data from previously collected genetic cohorts and electronic health record (EHR) data for 195 individuals with SLE. Single nucleotide polymorphisms (SNPs) were typed on the ImmunoChip. We included 95 variants previously associated with SLE risk. Variables extracted from the EHR included age, sex, race, ethnicity, age at diagnosis, and disease-associated laboratories: complement C3 and C4, SSA, SSB, RNP, anti-Smith, and anti-dsDNA. We first performed K-means clustering using all lab measures on the top N eigenvectors from principal component analysis determined using a bootstrap resampling strategy. We then used Chi-squared and ANOVA tests to examine whether demographic, clinical, and genetic variables were associated with each cluster.
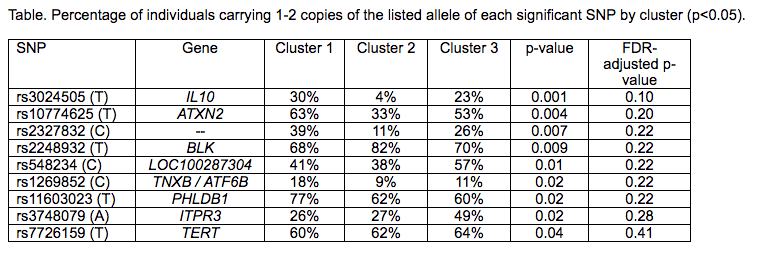
Results: 91% of patients were female; 47% were white, 13% African-American, 15% Asian, and 24% other/mixed race. Results demonstrated three distinct stable clusters (stability score >0.80) (Figure). Cluster 1 (n=91) was characterized as predominately white, non-Hispanic/Latino patients with higher age of onset (p<0.05). Cluster 2 (n=47) was significantly more likely to have + anti-dsDNA, + SSB, + RNP, and + anti-Sm antibodies (p<0.05). Cluster 3 (n=55) had a significantly higher percentage of abnormal C3 and C4 levels, + SSA, and lower age of onset (p<0.05). Nine SNPs were associated with the clusters but did not remain significant after multiple testing correction (Table).
Conclusion: Unsupervised clustering using sociodemographic and clinical variables derived from the EHR and genetic data identified three distinct subgroups of individuals with SLE. Future work will further define these genotype-phenotype clusters and perform validation studies in additional cohorts with more statistical power. Our findings may assist in identifying disease treatments for SLE using a more personalized approach.
To cite this abstract in AMA style:
Gianfrancesco M, Paranjpe I, Kay J, Nitiham J, Taylor K, Lanata C, Sirota M, Criswell LA, Schmajuk G, Yazdany J. Identification of Systemic Lupus Erythematosus Subgroups Using Electronic Health Record and Genetic Databases [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/identification-of-systemic-lupus-erythematosus-subgroups-using-electronic-health-record-and-genetic-databases/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-systemic-lupus-erythematosus-subgroups-using-electronic-health-record-and-genetic-databases/