Session Information
Session Type: ACR Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: The lack of a reproducible and quantitative method to accurately assess histological skin fibrosis in patients with systemic sclerosis (SSc) has long plagued clinical studies. Deep neural network (DNN) analysis, a computer vision technique, has the potential to radically augment histopathological analyses for myriad diseases including SSc. DNN analysis is a computational tool that extracts and quantifies complex image patterns. To test the hypothesis that DNN analysis applied to SSc skin biopsies is a useful method for dermal fibrosis quantification, the association between DNN outputs and skin score was determined.
Methods: One rheumatologist performed modified Rodnan skin score (mRSS) assessments followed by four mm dermal, punch biopsies of the non-dominant volar forearm, in patients with SSc. Biopsies were repeated at 6-, 12- 24-, and 36- months. Trichrome-stained sections were photomicrographed and transformed into quantitative features using AlexNet in the Matlab Neural Network Toolbox. Principal component analysis was used to identify the combination of quantitative features that captured the most quantitative feature variance. The correlation between the first principal component, mRSS, and local arm skin score was assessed.
Results: Twenty-six photomicrographs were analyzed. DNN analysis identified 4096 unique quantitative features. The first principal component of these 4096 quantitative features strongly correlated with local skin score (R = 0.52) and mRSS (R = 0.71).
Conclusion: This investigation demonstrates that DNN-derived quantitative features of SSc biopsies are sensitive to both local skin score and mRSS. Future directions include testing for generalizability in a larger sample and evaluating whether DNN are sensitive to biochemical measures of fibrosis. If validated, this suggests that DNN analysis can dramatically expand the quantifiable SSc phenome to permit inclusion of histological information into SSc trajectory models.
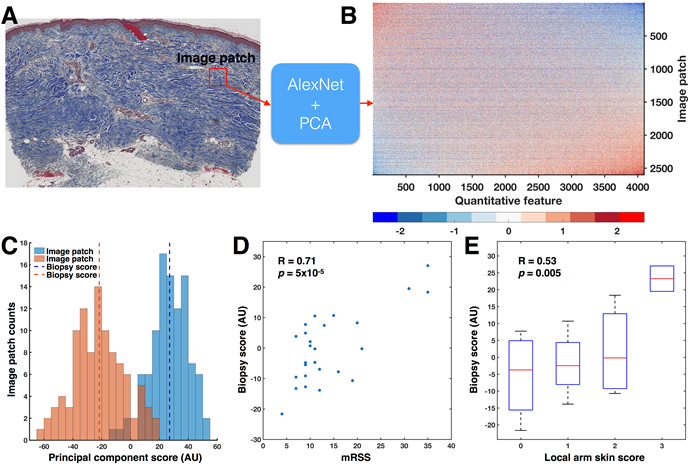
Figure 1:
A) 100 image patches (red square) were randomly sampled from the dermis and input to AlexNet. B) For each image patch, AlexNet identified 4096 quantitative features, resulting in a 2600 x 4096-dimensional matrix. The rows and columns of the quantitative feature matrix have been sorted by the first principal component (PC). C) Histograms of the principal component scores for image patches from the patients with the highest (35) and lowest (4) mRSS show little overlap. Across all 2600 image patches grouped by biopsy, the biopsy accounted for 30% of the total variance in the principal component scores (p < 10-100, one-way ANOVA). Therefore, biopsies were summarized into biopsy scores by taking the mean of the 100 patches. D) Biopsy score significantly correlated with mRSS (Pearson’s R = 0.71, p = 5×10-5). E) Biopsy score significantly correlated to local skin score (Pearson’s R = 0.53, p = 0.0049).
To cite this abstract in AMA style:
Correia C, Mawe S, Carns MA, Aren K, Hoffman A, Hinchcliff M, Mahoney JM. High-Throughput Quantitative Histology in Systemic Sclerosis Skin Disease Using Computer Vision [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/high-throughput-quantitative-histology-in-systemic-sclerosis-skin-disease-using-computer-vision/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/high-throughput-quantitative-histology-in-systemic-sclerosis-skin-disease-using-computer-vision/