Session Information
Session Type: ACR Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose:
In the clinical approach to CNS vasculitis, exclusion of infection is of major concern as some microbes can cause vasculitis, and infections can complicate immunosuppressive therapy for CNS vasculitis. The yields of conventional microbiologic techniques to detect CNS infections from cerebrospinal fluid (CSF) are suboptimal, particularly for unusual organisms that may not be part of the differential diagnosis. Unbiased metagenomic next-generation sequencing (mNGS) is an emerging technique that offers an alternative, hypothesis-free approach for detecting CNS infections. Here we report on a pilot study to examine the utility of mNGS in the management of patients with CNS vasculitis.
Methods:
Patients were recruited from our CNS registry at Cleveland Clinic. Total RNA was extracted from 250 uL of CSF, and cDNA sequencing libraries were prepared using random hexamer primers. NGS was performed on an Illumina HiSeq 2500 machine, and the resulting 10-20 million paired-end 150 nucleotide sequences were analyzed by a rapid computational pipeline for identifying all known viruses, bacteria, fungi and parasites. The patients received a regular medical care otherwise. The results of mNGS were compared with regular medical care.
Results:
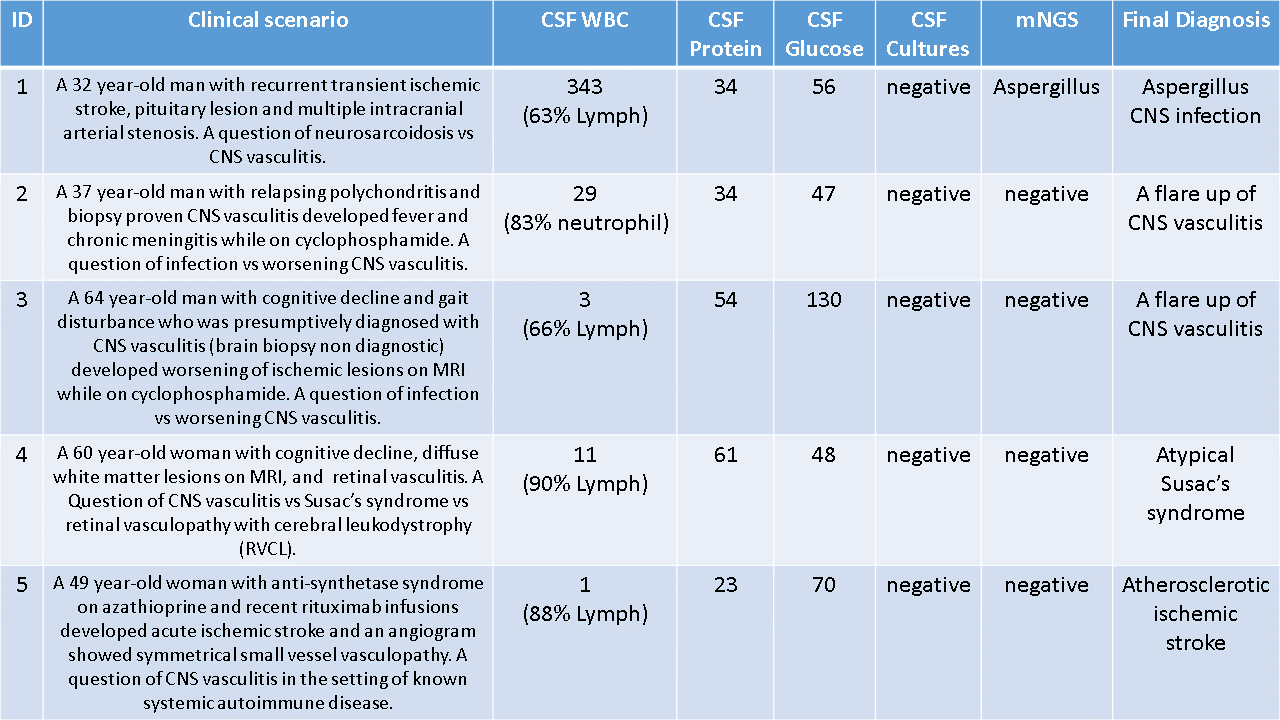
The main results are summarized in Table 1.
mNGS was useful in the following cases. Patient 1 was presumptively diagnosed with an autoimmune CNS disease based on a negative extensive infectious work-up. The CSF fungal cultures were negative 3 times, but research-based mNGS detected Aspergillus spp. This result was confirmed with a clinical 18s rRNA fungal PCR. Two patients (2, 3) with established CNS vasculitis clinically deteriorated while appropriately treated with glucocorticoids and cyclophosphamide. mNGS did not identify an infection. The patients were treated successfully with escalation of immunosuppression. Patient 4 did not have the typical triad for Susac’s syndrome, and as a result, a viral etiology was of great concern. To the degree that mNGS can be shown to have good sensitivity, the negative mNGS result in this case, would help exclude any lingering concerns about a potential infectious etiology.
Conclusion:
While these data are preliminary, mNGS may be useful to detect infectious pathogens that are novel or associated with a low yield with conventional diagnostic techniques in patients with CNS vasculitis. mNGS may be also useful to exclude opportunisitic infections in patients with CNS vasculitis arising in those treated with aggressive immunosuppression therapy. This technique is worth exploring in the larger cohort of patients with CNS vasculitis and patient recruitment is ongoing.
To cite this abstract in AMA style:
Tamaki H, Wilson MR, Calabrese LH, DeRisi JL, Hajj-Ali RA. The Utility of Unbiased Metagenomic Next Generation Sequencing in the Management of Patients with CNS Vasculitis [abstract]. Arthritis Rheumatol. 2017; 69 (suppl 10). https://acrabstracts.org/abstract/the-utility-of-unbiased-metagenomic-next-generation-sequencing-in-the-management-of-patients-with-cns-vasculitis/. Accessed .« Back to 2017 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/the-utility-of-unbiased-metagenomic-next-generation-sequencing-in-the-management-of-patients-with-cns-vasculitis/