Session Information
Session Type: ACR Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: According to current standards, methotrexate (MTX) is an anchor drug in the treatment of rheumatoid arthritis (RA) and should be used in the initial line of treatment in newly diagnosed patients. Some of these patients do not need additional therapy to reduce disease activity and even achieve sustained drug-free remission (sDFR) after tapering and stopping MTX. To identify these patients, we performed network analyses within differentially expressed genes (DEGs) and identified clusters (i.e. modules) of co-expressed genes associated with achieving sDFR.
Methods: Data was used from DMARD-naïve patients with early RA who in the U-Act-Early trial were randomized to initiate treat-to-target MTX therapy. MTX was given at a starting dose of 10 mg/week orally and was increased with 5 mg every 4 weeks until 30 mg or the maximum tolerable dose. When the treatment target, sustained remission (defined as disease activity score assessing 28 joints (DAS28) <2.6 with ≤4 swollen joints for ≥24 weeks), was achieved, therapy was tapered and hereafter stopped if remission was maintained. Patients achieved sDFR if they remained ≥3 months in remission while being-drug free. Blood samples were collected of those achieving sDFR (n=13) and those not able to discontinue medication (n=11) as controls. Hereafter ‘cluster of differentiation 4’-positive (CD4+) T Helper cells and CD14+ monocytes were isolated and analyzed using RNA sequencing. DEGs were identified and weighted gene co-expression network analysis was used to identify clusters (i.e. modules) of co-expressed genes.
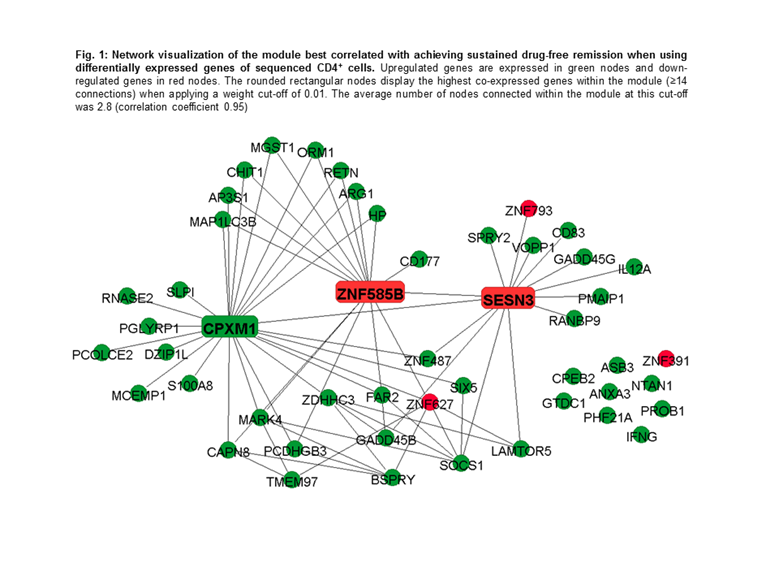
Results: Nine modules were identified in CD4+ cells and the module best correlated with achieving sDFR (Pearson correlation coefficient 0.60, p=0.012) included 49 co-expressed genes. Within this module, when performing pathway analyses in the Gene Ontology (GO) database, 304 terms were significantly overrepresented. Of these, response to bacterium (p=1.92E-07), response to external biotic stimulus (p=6.11E-07), and response to other organism (p=6.11E-07) were the most significant. In addition, two significant enriched pathways were found in the Kyoto Encyclopedia of Genes and Genomics database: “p53 signaling pathway” (p=8.44E-06) and “Jak-STAT signaling pathway” (p=2.22E-04). The down-regulated SESN3 and ZNF585B and the upregulated CPXM1 genes showed the highest intramodular connectivity and are therefore considered as signature genes (Fig. 1). Network analyses in CD14+ cells yielded no significant modules.
Conclusion: By network analyses of differentially expressed genes, several pathways were identified important for achieving sDFR in DMARD-naïve with early RA after initiation of an MTX-based strategy. SESN3, ZNF585B and CPXM1 were identified as signature genes that might be used as biomarkers for RA outcome.
To cite this abstract in AMA style:
Teitsma XM, Jacobs JW, Mokry M, Pethö-Schramm A, Borm ME, van Laar JM, Bijlsma JWJ, Lafeber FP. Differentially Co-Expressed Gene Networks in Previously DMARD-Naïve Patients with Early RA Achieving Sustained Drug-Free Remission after Step-up Methotrexate Therapy [abstract]. Arthritis Rheumatol. 2017; 69 (suppl 10). https://acrabstracts.org/abstract/differentially-co-expressed-gene-networks-in-previously-dmard-naive-patients-with-early-ra-achieving-sustained-drug-free-remission-after-step-up-methotrexate-therapy/. Accessed .« Back to 2017 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/differentially-co-expressed-gene-networks-in-previously-dmard-naive-patients-with-early-ra-achieving-sustained-drug-free-remission-after-step-up-methotrexate-therapy/