Session Information
Session Type: Abstract Submissions
Session Time: 5:30PM-7:00PM
Background/Purpose: Juvenile idiopathic arthritis (JIA) is a heterogeneous group of diseases which have in common inflammatory arthritis, but distinct clinical and genetic associations. Using biological information to accurately classify JIA into subtypes will enhance our understanding of the molecular pathogenesis of JIA and allow for personalized therapeutic options. Gene expression profiling allows inspection of the entire transcriptome for patterns of gene activity that can be used to interpret signals from genome-wide association studies and elucidate the pathways that contribute to distinct disease subtypes.
Methods: We performed RNA-Seq transcriptome profiling of peripheral blood samples from 129 children with JIA (45 oligoJIA; 54 polyJIA; 30 systemic sJIA), as well as 12 healthy controls. For an inflammatory disease outgroup comparison we also included 76 inflammatory bowel disease (IBD) samples (61 Crohn’s disease – CD; 15 ulcerative colitis – UC) samples. Pairwise differential expression analysis between cases and controls, and subtypes of disease was performed using edgeR software. We then extracted eigengenes representing 249 conserved Blood Transcript Modules (BTM), each capturing some aspect of immune gene expression, and characterized differences among phenotype categories.
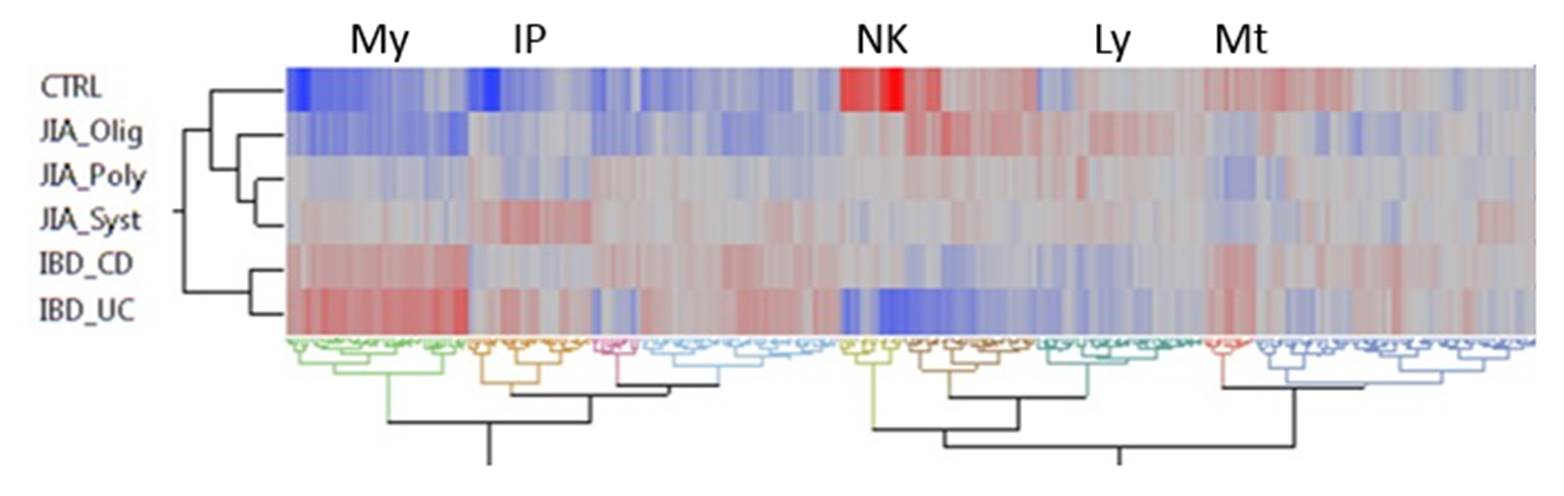
Results: Overall peripheral blood gene expression in JIA shows a gradient (see Figure) of disrupted gene expression involving hundreds of BTM, from healthy controls to oligo to polyarticular then sJIA. IBD most resembles sJIA, although with some key differences. Myeloid gene expression tends to be elevated in IBD (My), and lymphoid suppressed (Ly), with JIA intermediate. In addition, UC has a specific deficit in NK cell gene expression (NK), sJIA has a unique signature including inositol metabolism (IP), and JIA in general shows reduced mitochondrial gene activity Mt).
Conclusion: Transcriptomic analysis of whole peripheral blood identifies hundreds of modules that are specific to individual categories of disease. These may represent cell-specific modification of gene expression, or specific depletion or elevation of cells such as NK cells from the circulating blood following migration to the gut mucosa or joints. Ongoing studies are examining the influence of drug treatment on the profiles, and relating the differential expression to mechanisms of regulation of transcription.
To cite this abstract in AMA style:
Marigota U, Mo A, Prince J, Chan LHK, Kugathasan S, Gibson G, Prahalad S. Modular Gene Expression Discrimination of Juvenile Idiopathic Arthritis and Inflammatory Bowel Disease Subphenotypes in Peripheral Blood [abstract]. Arthritis Rheumatol. 2017; 69 (suppl 4). https://acrabstracts.org/abstract/modular-gene-expression-discrimination-of-juvenile-idiopathic-arthritis-and-inflammatory-bowel-disease-subphenotypes-in-peripheral-blood/. Accessed .« Back to 2017 Pediatric Rheumatology Symposium
ACR Meeting Abstracts - https://acrabstracts.org/abstract/modular-gene-expression-discrimination-of-juvenile-idiopathic-arthritis-and-inflammatory-bowel-disease-subphenotypes-in-peripheral-blood/