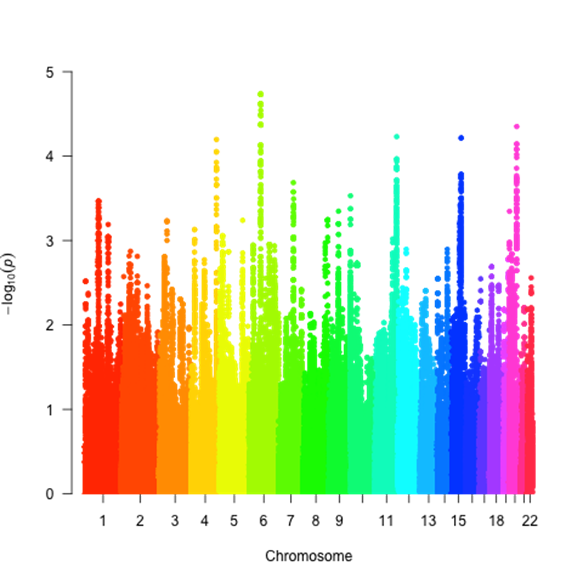Session Information
Session Type: ACR Concurrent Abstract Session
Session Time: 4:30PM-6:00PM
Background/Purpose: Juvenile idiopathic
arthritis (JIA) is a complex genetic trait. While several risk variants have
been identified using genome wide studies, a substantial component of the
genetic risk is unexplained by known variants. We hypothesized that distantly
related JIA cases might share genomic regions that harbor susceptibility
variants.
Methods: Using probabilistic linking of children with
JIA to the Utah Population Database, we identified distantly related JIA cases
among descendants of founders with 4 or more descendants affected with JIA. We
also identified distantly related controls whose degrees of relationship matched
our cases. We genotyped subjects using Affymetrix 6.0 microarrays, and after
stringent quality control, analyzed them using the Refined IBD program in
Beagle. We identified segments greater than 1000 basepairs shared pairwise
among cases. We
then used GraphIBD to test for single nucleotide polymorphisms (SNPs) that fell
within shared regions of case/case pairs more often control/control or
case/control pairs.
Results: We identified 296 distantly related cases and 300
distantly related controls. In all 194 founders had significantly excessive
number of descendants with JIA (range 4-17). We genotyped 218 cases and 148
controls, of whom 190 cases and 139 controls met our rigorous quality control inclusion
criteria (excluding individuals with <95% call rate or heterozygosity outliers,
close relatives, and population stratification outliers). Although no SNPs demonstrated
association at genome wide thresholds of significance, many demonstrated
associations at p <1X10-4 (Figure). Of the top 500 SNPs, 120 were
on chr 6, corresponding to a gene EYS, (best p <1.8X10-5) which
is implicated in retinitis pigmentosa, and acute uveitis associated with
ankylosing spondylitis. Variants on chr 20, corresponding to the gene PTPRT,
implicated in rheumatoid arthritis were also associated (p<8 X10-5).
Other regions of interest included NFKRB on chr 11, and two novel
transcripts on chr 4 and 15.
Conclusion: Using a novel approach, we found variants in EYS
and other loci that are shared more often among distantly related JIA case pairs,
although the modest sample size limited our power. Investigating distantly
related JIA case pairs offers a complementary approach to the search for genes
underlying JIA risk. Analysis stratifying subjects by presence of uveitis, and
replication of high priority variants in an independent cohort of JIA are
underway.
Acknowledgements: NIAMS (AR060893), The
Marcus Foundation Inc.
To cite this abstract in AMA style:
Prahalad S, Broadaway KA, Bohnsack JF, Dodd A, Hersh AO, Angeles-Han ST, Jang SR, Sudman M, Thompson SD, Zwick M, Conneely K, Epstein M. Identification of Shared Genomic Regions in Distantly Related Pairs of Cases with Juvenile Idiopathic Arthritis [abstract]. Arthritis Rheumatol. 2015; 67 (suppl 10). https://acrabstracts.org/abstract/identification-of-shared-genomic-regions-in-distantly-related-pairs-of-cases-with-juvenile-idiopathic-arthritis/. Accessed .« Back to 2015 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-shared-genomic-regions-in-distantly-related-pairs-of-cases-with-juvenile-idiopathic-arthritis/